3OOY
 
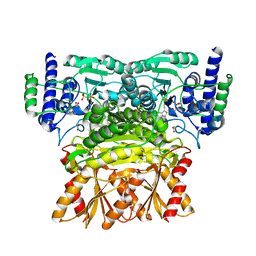 | | Crystal structure of human Transketolase (TKT) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Krojer, T, Krysztofinska, E, Guo, K, Pilka, E, Kochan, G, Chaikuad, A, Vollmar, M, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-31 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of human Transketolase (TKT)
To be Published
|
|
1BU6
 
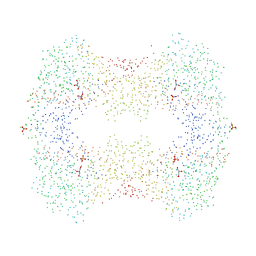 | | CRYSTAL STRUCTURES OF ESCHERICHIA COLI GLYCEROL KINASE AND THE MUTANT A65T IN AN INACTIVE TETRAMER: CONFORMATIONAL CHANGES AND IMPLICATIONS FOR ALLOSTERIC REGULATION | | Descriptor: | GLYCEROL, PROTEIN (GLYCEROL KINASE), SULFATE ION | | Authors: | Feese, M.D, Faber, H.R, Bystrom, C.E, Pettigrew, D.W, Remington, S.J. | | Deposit date: | 1998-08-30 | | Release date: | 1998-09-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Glycerol kinase from Escherichia coli and an Ala65-->Thr mutant: the crystal structures reveal conformational changes with implications for allosteric regulation.
Structure, 6, 1998
|
|
2WTJ
 
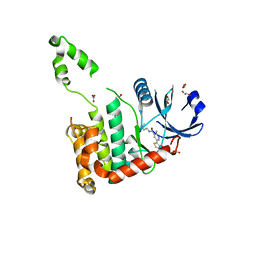 | | CRYSTAL STRUCTURE OF CHK2 IN COMPLEX WITH AN INHIBITOR | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5-(2,3-DIHYDROTHIENO[3,4-B][1,4]DIOXIN-5-YL)-N-[2-(DIMETHYLAMINO)ETHYL]PYRIDINE-3-CARBOXAMIDE, CHECKPOINT KINASE 2, ... | | Authors: | Hilton, S, Naud, S, Caldwell, J.J, Boxall, K, Burns, S, Anderson, V.E, Antoni, L, Allen, C.E, Pearl, L.H, Oliver, A.W, Aherne, G.W, Garrett, M.D, Collins, I. | | Deposit date: | 2009-09-16 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and Characterisation of 2-Aminopyridine Inhibitors of Checkpoint Kinase 2
Bioorg.Med.Chem., 18, 2010
|
|
1WU1
 
 | |
4QP9
 
 | |
3L2L
 
 | | X-ray Crystallographic Analysis of Pig Pancreatic Alpha-Amylase with Limit Dextrin and Oligosaccharide | | Descriptor: | CALCIUM ION, CHLORIDE ION, Pancreatic alpha-amylase, ... | | Authors: | Larson, S.B, Day, J.S, McPherson, A. | | Deposit date: | 2009-12-15 | | Release date: | 2010-04-14 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | X-ray crystallographic analyses of pig pancreatic alpha-amylase with limit dextrin, oligosaccharide, and alpha-cyclodextrin.
Biochemistry, 49, 2010
|
|
2RDK
 
 | | Five site mutated Cyanovirin-N with Mannose dimer bound | | Descriptor: | Cyanovirin-N, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Fromme, R, Katiliene, Z, Fromme, P, Ghirlanda, G. | | Deposit date: | 2007-09-24 | | Release date: | 2008-05-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conformational gating of dimannose binding to the antiviral protein cyanovirin revealed from the crystal structure at 1.35 A resolution.
Protein Sci., 17, 2008
|
|
3D02
 
 | |
3P1D
 
 | | Crystal structure of the bromodomain of human CREBBP in complex with N-Methyl-2-pyrrolidone (NMP) | | Descriptor: | 1-methylpyrrolidin-2-one, CREB-binding protein, POTASSIUM ION, ... | | Authors: | Filippakopoulos, P, Picaud, S, Feletar, I, Fedorov, O, Muniz, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-30 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
4Q9S
 
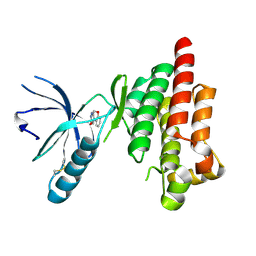 | | Crystal Structure of human Focal Adhesion Kinase (Fak) bound to Compound1 (3,5-DIHYDRO[1,2,4]TRIAZINO[3,4-C][1,4]BENZOXAZIN-2(1H)-ONE) | | Descriptor: | 3,5-dihydro[1,2,4]triazino[3,4-c][1,4]benzoxazin-2(1H)-one, Focal adhesion kinase 1 | | Authors: | Argiriadi, M.A, George, D.M. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of Selective and Orally Bioavailable Protein Kinase C theta (PKC theta ) Inhibitors from a Fragment Hit.
J.Med.Chem., 58, 2015
|
|
1C29
 
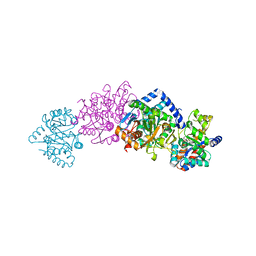 | | CRYSTAL STRUCTURE OF THE COMPLEX OF BACTERIAL TRYPTOPHAN SYNTHASE WITH THE TRANSITION STATE ANALOGUE INHIBITOR 4-(2-HYDROXYPHENYLTHIO)-1-BUTENYLPHOSPHONIC ACID | | Descriptor: | 4-(2-HYDROXYPHENYLTHIO)-1-BUTENYLPHOSPHONIC ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Sachpatzidis, A, Dealwis, C, Lubetsky, J.B, Liang, P.H, Anderson, K.S, Lolis, E. | | Deposit date: | 1999-07-23 | | Release date: | 2000-01-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic studies of phosphonate-based alpha-reaction transition-state analogues complexed to tryptophan synthase.
Biochemistry, 38, 1999
|
|
1K72
 
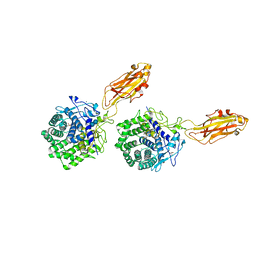 | | The X-ray Crystal Structure Of Cel9G Complexed With cellotriose | | Descriptor: | CALCIUM ION, Endoglucanase 9G, GLYCEROL, ... | | Authors: | Mandelman, D, Belaich, A, Belaich, J.P, Aghajari, N, Driguez, H, Haser, R. | | Deposit date: | 2001-10-18 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray Crystal Structure of the Multidomain Endoglucanase Cel9G from Clostridium
cellulolyticum Complexed with Natural and Synthetic Cello-Oligosaccharides
J.BACTERIOL., 185, 2003
|
|
5QJV
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z281802060 | | Descriptor: | 1,2-ETHANEDIOL, 4-methoxy-N-methylpyrimidin-2-amine, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
1NAP
 
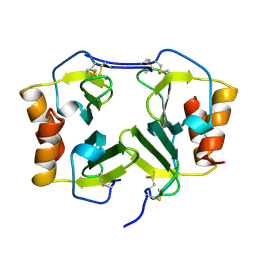 | |
5QJJ
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z24758179 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QJY
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z94597856 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
4DNO
 
 | | Crystal structure of the CFTR inhibitory factor Cif with the E153Q mutation adducted with the 1,2-epoxyhexane hydrolysis intermediate | | Descriptor: | (2R)-hexane-1,2-diol, Putative hydrolase | | Authors: | Bahl, C.D, Patankar, Y.R, Madden, D.R. | | Deposit date: | 2012-02-08 | | Release date: | 2013-08-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Visualizing the Mechanism of Epoxide Hydrolysis by the Bacterial Virulence Enzyme Cif.
Biochemistry, 55, 2016
|
|
5QOT
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with Z1592710382 | | Descriptor: | 1,2-ETHANEDIOL, 1-(difluoromethyl)-N-[(4-fluorophenyl)methyl]-1H-pyrazole-3-carboxamide, ACETATE ION, ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QP8
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with PB1787571279 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DCP2 (NUDT20), ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
1BXO
 
 | | ACID PROTEINASE (PENICILLOPEPSIN) (E.C.3.4.23.20) COMPLEX WITH PHOSPHONATE INHIBITOR: METHYL CYCLO[(2S)-2-[[(1R)-1-(N-(L-N-(3-METHYLBUTANOYL)VALYL-L-ASPARTYL)AMINO)-3-METHYLBUT YL] HYDROXYPHOSPHINYLOXY]-3-(3-AMINOMETHYL) PHENYLPROPANOATE | | Descriptor: | GLYCEROL, METHYL CYCLO[(2S)-2-[[(1R)-1-(N-(L-N-(3-METHYLBUTANOYL)VALYL-L-ASPARTYL)AMINO)-3-METHYLBUTYL]HYDROXYPHOSPHINYLOXY]-3-(3-AMINOMETHYL)PHENYLPROPANOATE, PROTEIN (PENICILLOPEPSIN), ... | | Authors: | Khan, A.R, Parrish, J.C, Fraser, M.E, Smith, W.W, Bartlett, P.A, James, M.N.G. | | Deposit date: | 1998-10-07 | | Release date: | 1998-10-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Lowering the entropic barrier for binding conformationally flexible inhibitors to enzymes.
Biochemistry, 37, 1998
|
|
3FVS
 
 | | Human Kynurenine Aminotransferase I in complex with Glycerol | | Descriptor: | GLYCEROL, Kynurenine--oxoglutarate transaminase 1, SODIUM ION | | Authors: | Han, Q, Robinson, H, Cai, T, Tagle, D.A, Li, J. | | Deposit date: | 2009-01-16 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insight into the inhibition of human kynurenine aminotransferase I/glutamine transaminase K
J.Med.Chem., 52, 2009
|
|
2XKK
 
 | | CRYSTAL STRUCTURE OF MOXIFLOXACIN, DNA, and A. BAUMANNII TOPO IV (PARE-PARC FUSION TRUNCATE) | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA, MAGNESIUM ION, ... | | Authors: | Wohlkonig, A, Chan, P.F, Fosberry, A.P, Homes, P, Huang, J, Kranz, M, Leydon, V.R, Miles, T.J, Pearson, N.D, Perera, R.L, Shillings, A.J, Gwynn, M.N, Bax, B.D. | | Deposit date: | 2010-07-08 | | Release date: | 2010-09-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural Basis of Quinolone Inhibition of Type Iia Topoisomerases and Target-Mediated Resistance
Nat.Struct.Mol.Biol., 17, 2010
|
|
3PA5
 
 | | X-ray crystal structure of compound 1 bound to human CHK1 kinase domain | | Descriptor: | 2-(carbamoylamino)-5-(4-chlorophenyl)-N-[(3S)-piperidin-3-yl]thiophene-3-carboxamide, GLYCEROL, Serine/threonine-protein kinase Chk1 | | Authors: | Fischmann, T.O. | | Deposit date: | 2010-10-18 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, synthesis and SAR of thienopyridines as potent CHK1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1C3Q
 
 | | CRYSTAL STRUCTURE OF NATIVE THIAZOLE KINASE IN THE MONOCLINIC FORM | | Descriptor: | 2-(4-METHYL-THIAZOL-5-YL)-ETHANOL, CHLORIDE ION, Hydroxyethylthiazole kinase | | Authors: | Campobasso, N, Mathews, I.I, Begley, T.P, Ealick, S.E. | | Deposit date: | 1999-07-28 | | Release date: | 1999-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 4-methyl-5-beta-hydroxyethylthiazole kinase from Bacillus subtilis at 1.5 A resolution.
Biochemistry, 39, 2000
|
|
5QP9
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with Z100435060 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DCP2 (NUDT20), ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
