6M6R
 
 | | Crystal structure of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3) C-terminal domain with 5'-ppp 8-mer ssRNA | | Descriptor: | Dicer Related Helicase, RNA (5'-R(*(GTP)P*GP*CP*CP*GP*CP*CP*C)-3'), ZINC ION | | Authors: | Li, K, Zheng, J, Wirawan, M, Xiong, Z, Fedorova, O, Griffin, P, Plyle, A, Luo, D. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Insights into the structure and RNA-binding specificity of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3).
Nucleic Acids Res., 49, 2021
|
|
6M6S
 
 | | Crystal structure of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3) C-terminal domain with 5'-ppp 12-mer dsRNA | | Descriptor: | Dicer Related Helicase, RNA (5'-R(*(GTP)P*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*C)-3'), ZINC ION | | Authors: | Li, K, Zheng, J, Wirawan, M, Xiong, Z, Fedorova, O, Griffin, P, Plyle, A, Luo, D. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into the structure and RNA-binding specificity of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3).
Nucleic Acids Res., 49, 2021
|
|
7SWF
 
 | |
3AL0
 
 | | Crystal structure of the glutamine transamidosome from Thermotoga maritima in the glutamylation state. | | Descriptor: | Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, Glutamyl-tRNA(Gln) amidotransferase subunit A, Glutamyl-tRNA(Gln) amidotransferase subunit C,Linker,Glutamate--tRNA ligase 2, ... | | Authors: | Ito, T, Yokoyama, S. | | Deposit date: | 2010-07-19 | | Release date: | 2010-09-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.368 Å) | | Cite: | Two enzymes bound to one transfer RNA assume alternative conformations for consecutive reactions.
Nature, 467, 2010
|
|
5I61
 
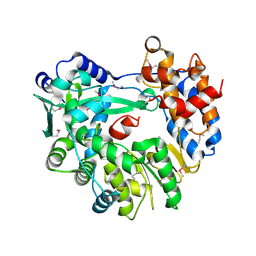 | |
6UV1
 
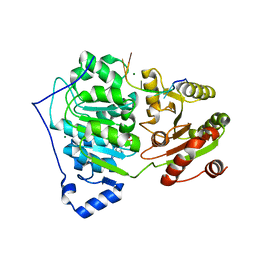 | | Crystal structure of RNA helicase DDX17 in complex of rU10 RNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Ngo, T.D, Partin, A.C, Nam, Y. | | Deposit date: | 2019-11-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | RNA Specificity and Autoregulation of DDX17, a Modulator of MicroRNA Biogenesis.
Cell Rep, 29, 2019
|
|
6UV2
 
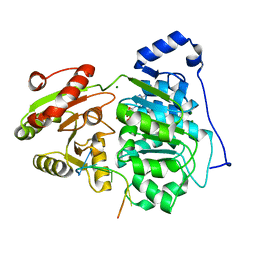 | | Crystal structure of the core domain of RNA helicase DDX17 with RNA pri-125a-oligo1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Ngo, T.D, Partin, A.C, Nam, Y. | | Deposit date: | 2019-11-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | RNA Specificity and Autoregulation of DDX17, a Modulator of MicroRNA Biogenesis.
Cell Rep, 29, 2019
|
|
5I2T
 
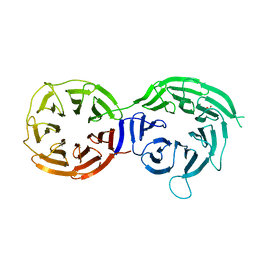 | |
6OL3
 
 | | Crystal structure of an adenovirus virus-associated RNA | | Descriptor: | Adenovirus Virus-Associated (VA) RNA I apical and central domains, POTASSIUM ION | | Authors: | Hood, I.V, Gordon, J.M, Bou-Nader, C, Henderson, F.V, Bahmanjah, S, Zhang, J. | | Deposit date: | 2019-04-15 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal structure of an adenovirus virus-associated RNA.
Nat Commun, 10, 2019
|
|
4YOE
 
 | | Structure of UP1 bound to RNA 5'-AGU-3' | | Descriptor: | ACETATE ION, Heterogeneous nuclear ribonucleoprotein A1, RNA AGU, ... | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2015-03-11 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The First Crystal Structure of the UP1 Domain of hnRNP A1 Bound to RNA Reveals a New Look for an Old RNA Binding Protein.
J.Mol.Biol., 427, 2015
|
|
7THM
 
 | | SARS-CoV-2 nsp12/7/8 complex with a native N-terminus nsp9 | | Descriptor: | MANGANESE (II) ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Osinski, A, Tagliabracci, V.S, Chen, Z, Li, Y. | | Deposit date: | 2022-01-11 | | Release date: | 2022-03-16 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | The mechanism of RNA capping by SARS-CoV-2.
Nature, 609, 2022
|
|
6KF3
 
 | | Cryo-EM structure of Thermococcus kodakarensis RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit A'', DNA-directed RNA polymerase subunit D, ... | | Authors: | Jun, S.-H, Hyun, J, Jeong, H, Cha, J.S, Kim, H, Bartlett, M.S, Cho, H.-S, Murakami, K.S. | | Deposit date: | 2019-07-06 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Direct binding of TFE alpha opens DNA binding cleft of RNA polymerase.
Nat Commun, 11, 2020
|
|
6KF9
 
 | | Cryo-EM structure of Thermococcus kodakarensis RNA polymerase | | Descriptor: | DNA (27-MER), DNA (5'-D(P*TP*CP*GP*GP*TP*AP*AP*TP*CP*AP*CP*GP*CP*TP*CP*C)-3'), DNA-directed RNA polymerase subunit, ... | | Authors: | Jun, S.-H, Hyun, J, Jeong, H, Cha, J.S, Kim, H, Bartlett, M.S, Cho, H.-S, Murakami, K.S. | | Deposit date: | 2019-07-07 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Direct binding of TFE alpha opens DNA binding cleft of RNA polymerase.
Nat Commun, 11, 2020
|
|
5SB8
 
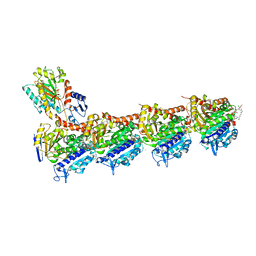 | | Tubulin-maytansinoid-3-complex | | Descriptor: | (1S,2R,3S,5S,6S,16E,18E,20R)-11-chloro-6-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18,21-hexaene-8,23-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Marzullo, P, Boiarska, Z, Perez-Pena, H, Abel, A.-C, Alvarez-Bernad, B, Lucena-Agell, D, Vasile, F, Sironi, M, Steinmetz, M.O, Prota, A.E, Diaz, J.F, Pieraccini, S, Passarella, D. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Maytansinol Derivatives: Side Reactions as a Chance for New Tubulin Binders.
Chemistry, 28, 2022
|
|
5SBD
 
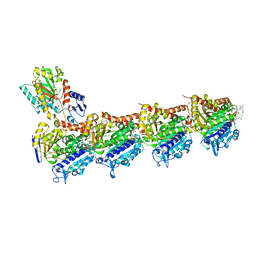 | | Tubulin-maytansinoid-5b-complex | | Descriptor: | (1S,2R,3S,5S,6S,16E,18E,20R)-11-chloro-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18,21(25)-hexaen-6-yl acetate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Marzullo, P, Boiarska, Z, Perez-Pena, H, Abel, A.-C, Alvarez-Bernad, B, Lucena-Agell, D, Vasile, F, Sironi, M, Steinmetz, M.O, Prota, A.E, Diaz, J.F, Pieraccini, S, Passarella, D. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Maytansinol Derivatives: Side Reactions as a Chance for New Tubulin Binders.
Chemistry, 28, 2022
|
|
5SBA
 
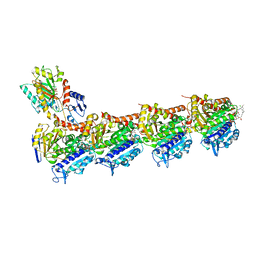 | | Tubulin-maytansinoid-4b-complex | | Descriptor: | (1S,2R,3S,5S,6S,16E,18E,20R,21S)-11-chloro-21-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaen-6-yl acetate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Marzullo, P, Boiarska, Z, Perez-Pena, H, Abel, A.-C, Alvarez-Bernad, B, Lucena-Agell, D, Vasile, F, Sironi, M, Steinmetz, M.O, Prota, A.E, Diaz, J.F, Pieraccini, S, Passarella, D. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Maytansinol Derivatives: Side Reactions as a Chance for New Tubulin Binders.
Chemistry, 28, 2022
|
|
5SBE
 
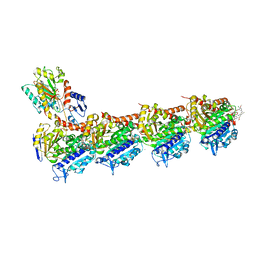 | | Tubulin-maytansinoid-5c-complex | | Descriptor: | (1S,2R,3S,5S,6S,16E,18E,20R)-11-chloro-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18,21-hexaen-6-yl hept-6-ynoate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Marzullo, P, Boiarska, Z, Perez-Pena, H, Abel, A.-C, Alvarez-Bernad, B, Lucena-Agell, D, Vasile, F, Sironi, M, Steinmetz, M.O, Prota, A.E, Diaz, J.F, Pieraccini, S, Passarella, D. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Maytansinol Derivatives: Side Reactions as a Chance for New Tubulin Binders.
Chemistry, 28, 2022
|
|
5SBB
 
 | | Tubulin-maytansinoid-4c-complex | | Descriptor: | (1R,2R,3S,5S,6S,16E,18E,20R,21S)-11-chloro-21-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaen-6-yl heptanoate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Marzullo, P, Boiarska, Z, Perez-Pena, H, Abel, A.-C, Alvarez-Bernad, B, Lucena-Agell, D, Vasile, F, Sironi, M, Steinmetz, M.O, Prota, A.E, Diaz, J.F, Pieraccini, S, Passarella, D. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Maytansinol Derivatives: Side Reactions as a Chance for New Tubulin Binders.
Chemistry, 28, 2022
|
|
5SBC
 
 | | Tubulin-maytansinoid-5a-complex | | Descriptor: | (1S,2R,3S,5S,6S,16E,18E,20R)-11-chloro-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18,21-hexaen-6-yl phenylacetate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Marzullo, P, Boiarska, Z, Perez-Pena, H, Abel, A.-C, Alvarez-Bernad, B, Lucena-Agell, D, Vasile, F, Sironi, M, Steinmetz, M.O, Prota, A.E, Diaz, J.F, Pieraccini, S, Passarella, D. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Maytansinol Derivatives: Side Reactions as a Chance for New Tubulin Binders.
Chemistry, 28, 2022
|
|
5SB9
 
 | | Tubulin-maytansinoid-4a-complex | | Descriptor: | (1S,2R,3S,5S,6S,16E,18E,20R,21S)-11-chloro-21-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaen-6-yl phenylacetate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Marzullo, P, Boiarska, Z, Perez-Pena, H, Abel, A.-C, Alvarez-Bernad, B, Lucena-Agell, D, Vasile, F, Sironi, M, Steinmetz, M.O, Prota, A.E, Diaz, J.F, Pieraccini, S, Passarella, D. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Maytansinol Derivatives: Side Reactions as a Chance for New Tubulin Binders.
Chemistry, 28, 2022
|
|
6KF4
 
 | | Cryo-EM structure of Thermococcus kodakarensis RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit A'', DNA-directed RNA polymerase subunit D, ... | | Authors: | Jun, S.-H, Hyun, J, Jeong, H, Cha, J.S, Kim, H, Bartlett, M.S, Cho, H.-S, Murakami, K.S. | | Deposit date: | 2019-07-06 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Direct binding of TFE alpha opens DNA binding cleft of RNA polymerase.
Nat Commun, 11, 2020
|
|
2TRA
 
 | | RESTRAINED REFINEMENT OF TWO CRYSTALLINE FORMS OF YEAST ASPARTIC ACID AND PHENYLALANINE TRANSFER RNA CRYSTALS | | Descriptor: | MAGNESIUM ION, SPERMINE, TRNAASP | | Authors: | Westhof, E, Dumas, P, Moras, D. | | Deposit date: | 1987-11-06 | | Release date: | 1987-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Restrained refinement of two crystalline forms of yeast aspartic acid and phenylalanine transfer RNA crystals.
Acta Crystallogr.,Sect.A, 44, 1988
|
|
6UV4
 
 | | Crystal structure of the core domain of RNA helicase DDX17 with RNA pri-18a-oligo1 | | Descriptor: | 18a_oligo1, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Ngo, T.D, Partin, A.C, Nam, Y. | | Deposit date: | 2019-11-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | RNA Specificity and Autoregulation of DDX17, a Modulator of MicroRNA Biogenesis.
Cell Rep, 29, 2019
|
|
6UV3
 
 | | Crystal structure of the core domain of RNA helicase DDX17 with RNA pri-125a-oligo2 | | Descriptor: | 125a-oligo2, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Ngo, T.D, Partin, A.C, Nam, Y. | | Deposit date: | 2019-11-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | RNA Specificity and Autoregulation of DDX17, a Modulator of MicroRNA Biogenesis.
Cell Rep, 29, 2019
|
|
6G63
 
 | | RNase E in complex with sRNA RrpA | | Descriptor: | RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3'), Ribonuclease E, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Bandyra, K.B, Luisi, B.F. | | Deposit date: | 2018-03-31 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Substrate Recognition and Autoinhibition in the Central Ribonuclease RNase E.
Mol. Cell, 72, 2018
|
|
