7Y38
 
 | |
1Q65
 
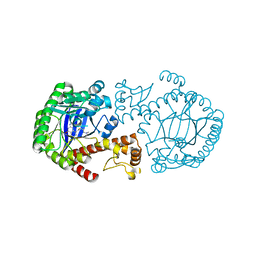 | | CRYSTAL STRUCTURE OF TGT IN COMPLEX WITH 2,6-DIAMINO-8-(2-dimethylaminoethylsulfanylmethyl)-3H-QUINAZOLIN-4-ONE crystallized at pH 5.5 | | Descriptor: | 2,6-DIAMINO-8-(2-DIMETHYLAMINOETHYLSULFANYLMETHYL)-3H-QUINAZOLIN-4-ONE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Meyer, E, Reuter, K, Stubbs, M.T, Garcia, G.A, Klebe, G. | | Deposit date: | 2003-08-12 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic Study of Inhibitors of tRNA-guanine Transglycosylase Suggests a New Structure-based Pharmacophore for Virtual Screening.
J.Mol.Biol., 338, 2004
|
|
1PSU
 
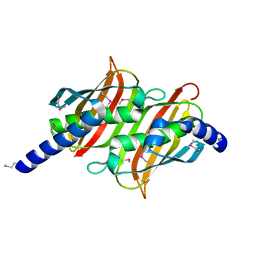 | | Structure of the E. coli PaaI protein from the phyenylacetic acid degradation operon | | Descriptor: | Phenylacetic acid degradation protein PaaI | | Authors: | Kniewel, R, Buglino, J, Solorzano, V, Wu, J, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-06-21 | | Release date: | 2003-07-08 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure, function, and mechanism of the phenylacetate pathway hot dog-fold thioesterase PaaI.
J.Biol.Chem., 281, 2006
|
|
1PUI
 
 | |
6FGQ
 
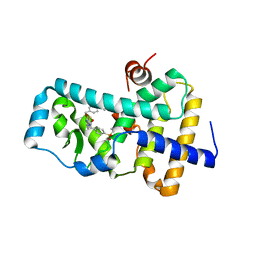 | | Ligand complex of RORg LBD | | Descriptor: | Nuclear receptor ROR-gamma, methyl 4-[[3-[5-[2-(4-ethylsulfonylphenyl)ethanoylamino]thiophen-3-yl]pyridin-2-yl]oxymethyl]benzoate | | Authors: | Xue, Y, Aagaard, A, Narjes, F. | | Deposit date: | 2018-01-11 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Potent and Orally Bioavailable Inverse Agonists of ROR gamma t Resulting from Structure-Based Design.
J. Med. Chem., 61, 2018
|
|
1PSQ
 
 | | Structure of a probable thiol peroxidase from Streptococcus pneumoniae | | Descriptor: | probable thiol peroxidase | | Authors: | Kniewel, R, Buglino, J, Solorzano, V, Wu, J, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-06-21 | | Release date: | 2003-07-08 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a probable thiol peroxidase from Streptococcus pneumoniae
To be Published
|
|
2BU1
 
 | | MS2-RNA HAIRPIN (5BRU -5) COMPLEX | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP *5BU*AP*CP*CP*CP*AP*UP*GP*U)-3', MS2 COAT PROTEIN | | Authors: | Grahn, E, Moss, T, Helgstrand, C, Fridborg, K, Sundaram, M, Tars, K, Lago, H, Stonehouse, N.J, Davis, D.R, Stockley, P.G, Liljas, L. | | Deposit date: | 2005-06-08 | | Release date: | 2005-08-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of pyrimidine specificity in the MS2 RNA hairpin-coat-protein complex.
Rna, 7, 2001
|
|
6FHI
 
 | |
1PWZ
 
 | | Crystal structure of the haloalcohol dehalogenase HheC complexed with (R)-styrene oxide and chloride | | Descriptor: | CHLORIDE ION, R-STYRENE OXIDE, halohydrin dehalogenase | | Authors: | de Jong, R.M, Tiesinga, J.J.W, Rozeboom, H.J, Kalk, K.H, Tang, L, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2003-07-02 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Mechanism of a Bacterial Haloalcohol Dehalogenase: a new variation of the short-chain dehydrogenase/reductase fold without an NAD(P)H binding site
EMBO J., 22, 2003
|
|
6DGP
 
 | |
1Q63
 
 | | CRYSTAL STRUCTURE OF TGT IN COMPLEX WITH 2,6-Diamino-8-(1H-imidazol-2-ylsulfanylmethyl)-3H-quinazoline-4-one crystallized at pH 5.5 | | Descriptor: | 2,6-DIAMINO-8-(1H-IMIDAZOL-2-YLSULFANYLMETHYL)-3H-QUINAZOLINE-4-ONE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Meyer, E, Reuter, K, Stubbs, M.T, Garcia, G.A, Klebe, G. | | Deposit date: | 2003-08-12 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic Study of Inhibitors of tRNA-guanine Transglycosylase Suggests a New Structure-based Pharmacophore for Virtual Screening.
J.Mol.Biol., 338, 2004
|
|
1ZUX
 
 | | EosFP Fluorescent Protein- Green Form | | Descriptor: | green to red photoconvertible GPF-like protein EosFP | | Authors: | Nar, H, Nienhaus, K, Wiedenmann, J, Nienhaus, G.U. | | Deposit date: | 2005-06-01 | | Release date: | 2005-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for photo-induced protein cleavage and green-to-red conversion of fluorescent protein EosFP.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
7XWZ
 
 | | Crystal structure of SARS-CoV-2 N-NTD and dsRNA complex | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Nucleoprotein, ... | | Authors: | Luan, X.D, Li, X.M, Li, Y.F. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Antiviral drug design based on structural insights into the N-terminal domain and C-terminal domain of the SARS-CoV-2 nucleocapsid protein.
Sci Bull (Beijing), 67, 2022
|
|
1RZN
 
 | |
1S3N
 
 | | Structural and Functional Characterization of a Novel Archaeal Phosphodiesterase | | Descriptor: | Hypothetical protein MJ0936, MANGANESE (II) ION | | Authors: | Chen, S, Busso, D, Yakunin, A.F, Kuznetsova, E, Proudfoot, M, Jancrick, J, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-01-13 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional characterization of a novel phosphodiesterase from Methanococcus jannaschii
J.Biol.Chem., 279, 2004
|
|
6DGQ
 
 | | Crystal Structure of Human PPARgamma Ligand Binding Domain in Complex with CAY10506 | | Descriptor: | N-(2-{4-[(2,4-dioxo-3,4-dihydro-2H-1lambda~4~,3-thiazol-5-yl)methyl]phenoxy}ethyl)-5-[(3R)-1,2-dithiolan-3-yl]pentanamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Shang, J, Kojetin, D.J. | | Deposit date: | 2018-05-17 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Quantitative structural assessment of graded receptor agonism.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7Y62
 
 | | Crystal structure of human TFEB HLHLZ domain | | Descriptor: | Transcription factor EB | | Authors: | Yang, G, Li, P, Lin, Y, Liu, Z, Sun, H, Zhao, Z, Fang, P, Wang, J. | | Deposit date: | 2022-06-18 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A small-molecule drug inhibits autophagy gene expression through the central regulator TFEB.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1SDS
 
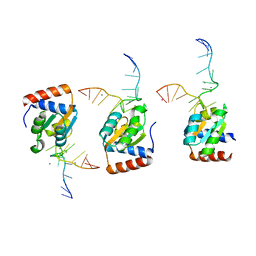 | |
6FZU
 
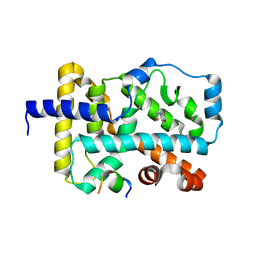 | |
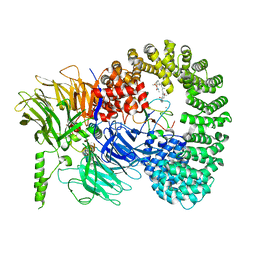
7B92
 
 | |
1WBO
 
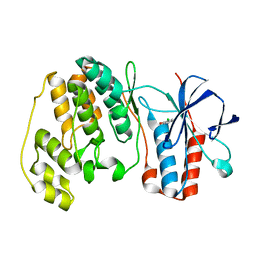 | | fragment based p38 inhibitors | | Descriptor: | 2-CHLOROPHENOL, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Cleasby, A, Devine, L, Gill, A, Jhoti, H. | | Deposit date: | 2004-11-04 | | Release date: | 2005-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Fragment-Based Lead Discovery Using X-Ray Crystallography
J.Med.Chem., 48, 2005
|
|
1WBW
 
 | | Identification of novel p38 alpha MAP Kinase inhibitors using fragment-based lead generation. | | Descriptor: | 3-(1-NAPHTHYLMETHOXY)PYRIDIN-2-AMINE, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Tickle, J, Cleasby, A, Devine, L.A, Jhoti, H. | | Deposit date: | 2004-11-05 | | Release date: | 2005-11-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Identification of Novel P38Alpha Map Kinase Inhibitors Using Fragment-Based Lead Generation
J.Med.Chem., 48, 2005
|
|
7B9C
 
 | |
7BO6
 
 | | VDR complex with LCA derivative | | Descriptor: | (4R)-4-[(3R,5R,8R,9S,10S,13R,14S,17R)-10,13-dimethyl-3-(2-methyl-2-oxidanyl-propyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]pentanoic acid, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2021-01-24 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Lithocholic acid-based design of noncalcemic vitamin D receptor agonists.
Bioorg.Chem., 111, 2021
|
|
7B91
 
 | |
