6U5P
 
 | | Calcium-bound MthK gating ring state 1 | | Descriptor: | CALCIUM ION, Calcium-gated potassium channel MthK | | Authors: | Fan, C, Nimigean, C.M. | | Deposit date: | 2019-08-28 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ball-and-chain inactivation in a calcium-gated potassium channel.
Nature, 580, 2020
|
|
7CAI
 
 | | SARS-CoV-2 S trimer with two RBDs in the open state and complexed with two H014 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H014 Fab, ... | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Wang, Y, Rao, Z, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
6X7F
 
 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B2 (TTC-B2) containing an mRNA with a 24 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
8VK4
 
 | |
8V9J
 
 | | Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Msmeg1130 (Balon) (Structure 4) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S14, ... | | Authors: | Rybak, M.Y, Helena-Bueno, K, Hill, C.H, Melnikov, S.V, Gagnon, M.G. | | Deposit date: | 2023-12-08 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A new family of bacterial ribosome hibernation factors.
Nature, 626, 2024
|
|
8VKM
 
 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (conformation 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
7CHH
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with BD-368-2 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BD-368-2 Fab heavy chain, ... | | Authors: | Xiao, J, Zhu, Q, Wang, G. | | Deposit date: | 2020-07-05 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structurally Resolved SARS-CoV-2 Antibody Shows High Efficacy in Severely Infected Hamsters and Provides a Potent Cocktail Pairing Strategy.
Cell, 183, 2020
|
|
8VKO
 
 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
7YRO
 
 | | Crystal structure of mango fucosyltransferase 13 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, Fucosyltransferase, ... | | Authors: | Okada, T, Teramoto, T, Ihara, H, Ikeda, Y, Kakuta, Y. | | Deposit date: | 2022-08-10 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structure of mango alpha 1,3/ alpha 1,4-fucosyltransferase elucidates unique elements that regulate Lewis A-dominant oligosaccharide assembly.
Glycobiology, 34, 2024
|
|
8V9K
 
 | | Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Rv2629 (Balon) (Structure 5) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S2, ... | | Authors: | Rybak, M.Y, Helena-Bueno, K, Hill, C.H, Melnikov, S.V, Gagnon, M.G. | | Deposit date: | 2023-12-08 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A new family of bacterial ribosome hibernation factors.
Nature, 626, 2024
|
|
7YMM
 
 | | PSII-Pcb Tetramer of Acaryochloris Marina | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Shen, L.L, Gao, Y.Z, Wang, W.D, Zhang, X, Shen, J.R, Wang, P.Y, Han, G.Y. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a unique PSII-Pcb tetrameric megacomplex in a chlorophyll d -containing cyanobacterium.
Sci Adv, 10, 2024
|
|
7YMI
 
 | | PSII-Pcb Dimer of Acaryochloris Marina | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Shen, L.L, Gao, Y.Z, Wang, W.D, Zhang, X, Shen, J.R, Wang, P.Y, Han, G.Y. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a unique PSII-Pcb tetrameric megacomplex in a chlorophyll d -containing cyanobacterium.
Sci Adv, 10, 2024
|
|
8VKL
 
 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (conformation 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
6X80
 
 | | Structure of the Campylobacter jejuni G508A Flagellar Filament | | Descriptor: | 5,7-diamino-3,5,7,9-tetradeoxy-L-glycero-alpha-L-manno-non-2-ulopyranosonic acid, Flagellin A | | Authors: | Kreutzberger, M.A.B, Wang, F, Egelman, E.H. | | Deposit date: | 2020-06-01 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Atomic structure of the Campylobacter jejuni flagellar filament reveals how epsilon Proteobacteria escaped Toll-like receptor 5 surveillance.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8V9L
 
 | | Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Msmeg1130 (Balon) and MsmegEF-Tu(GDP) (Composite structure 6) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S2, ... | | Authors: | Rybak, M.Y, Helena-Bueno, K, Hill, C.H, Melnikov, S.V, Gagnon, M.G. | | Deposit date: | 2023-12-08 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A new family of bacterial ribosome hibernation factors.
Nature, 626, 2024
|
|
6TXR
 
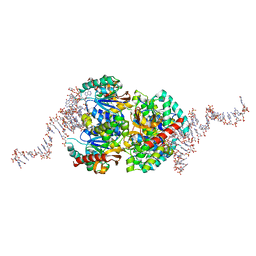 | | Structural insights into cubane-modified aptamer recognition of a malaria biomarker | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Cheung, Y, Roethlisberger, P, Mechaly, A, Weber, P, Wong, A, Lo, Y, Haouz, A, Savage, P, Hollenstein, M, Tanner, J. | | Deposit date: | 2020-01-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evolution of abiotic cubane chemistries in a nucleic acid aptamer allows selective recognition of a malaria biomarker.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6XM0
 
 | | Consensus structure of SARS-CoV-2 spike at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6U9O
 
 | | Human IMPDH2 treated with ATP, IMP, NAD+, and 2 mM GTP. Fully compressed filament segment reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-09 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6UB9
 
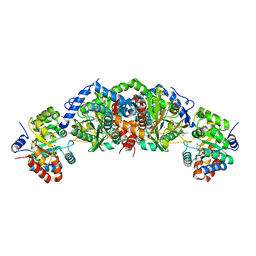 | | Crystal structure of tryptophan synthase from M. tuberculosis - AMINOACRYLATE- AND BRD6309-BOUND FORM | | Descriptor: | (2R,3S,4R)-3-(4'-chloro-2',6'-difluoro[1,1'-biphenyl]-4-yl)-4-(fluoromethyl)azetidine-2-carbonitrile, 1,2-ETHANEDIOL, 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, ... | | Authors: | Chang, C, Michalska, K, Maltseva, N.I, Jedrzejczak, R, McCarren, P, Nag, P.P, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-11 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.783 Å) | | Cite: | Crystal structure of tryptophan synthase from M. tuberculosis - AMINOACRYLATE- AND BRD6309-BOUND FORM
To be Published
|
|
8V80
 
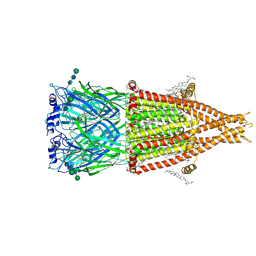 | | Alpha7-nicotinic acetylcholine receptor bound to epibatidine and (-)-TQS | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (3aR,4S,9bS)-4-(naphthalen-1-yl)-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-8-sulfonamide, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, ... | | Authors: | Burke, S.M, Noviello, C.M, Hibbs, R.E. | | Deposit date: | 2023-12-04 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Structural mechanisms of alpha 7 nicotinic receptor allosteric modulation and activation.
Cell, 187, 2024
|
|
6X6P
 
 | | Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Herrera, N.G, Morano, N.C, Celikgil, A, Georgiev, G.I, Malonis, R, Lee, J.H, Tong, K, Vergnolle, O, Massimi, A, Yen, L.Y, Noble, A.J, Kopylov, M, Bonanno, J.B, Garrett-Thompson, S.C, Hayes, D.B, Brenowitz, M, Garforth, S.J, Eng, E.T, Lai, J.R, Almo, S.C. | | Deposit date: | 2020-05-28 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis.
Biorxiv, 2020
|
|
8V8A
 
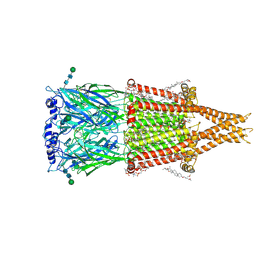 | | Alpha7-nicotinic acetylcholine receptor time resolved bound to epibatidine and PNU-120596 desensitized intermediate state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Burke, S.M, Noviello, C.M, Hibbs, R.E. | | Deposit date: | 2023-12-05 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Structural mechanisms of alpha 7 nicotinic receptor allosteric modulation and activation.
Cell, 187, 2024
|
|
8V89
 
 | | Alpha7-nicotinic acetylcholine receptor time resolved resting state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Burke, S.M, Noviello, C.M, Hibbs, R.E. | | Deposit date: | 2023-12-05 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural mechanisms of alpha 7 nicotinic receptor allosteric modulation and activation.
Cell, 187, 2024
|
|
6TZU
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, N84A mutant with pyruvate bound in the active site | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Majdi Yazdi, M, Lehnert, C, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2019-08-13 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Asparagine-84, a regulatory allosteric site residue, helps maintain the quaternary structure of Campylobacter jejuni dihydrodipicolinate synthase.
J.Struct.Biol., 209, 2020
|
|
8VKK
 
 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
