2R3X
 
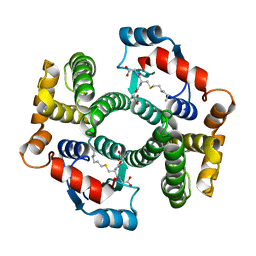 | | Crystal structure of an R15L hGSTA1-1 mutant complexed with S-hexyl-glutathione | | Descriptor: | Glutathione S-transferase A1, S-HEXYLGLUTATHIONE | | Authors: | Burke, J.P.W.G, Kinsley, N, Sayed, M, Sewell, T, Dirr, H.W. | | Deposit date: | 2007-08-30 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Arginine 15 stabilizes an S(N)Ar reaction transition state and the binding of anionic ligands at the active site of human glutathione transferase A1-1.
Biophys.Chem., 146, 2010
|
|
3E99
 
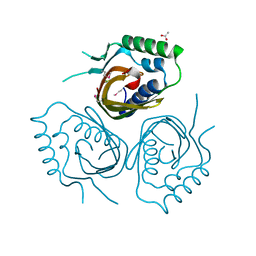 | |
4RTC
 
 | | Crystal structure of the green fluorescent variant, nowGFP, of the cyan Cerulean at pH 9.0 | | Descriptor: | GLYCEROL, nowGFP | | Authors: | Pletnev, V.Z, Pletneva, N.V, Pletnev, S.V. | | Deposit date: | 2014-11-14 | | Release date: | 2015-09-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of the green fluorescent protein NowGFP with an anionic tryptophan-based chromophore.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3D47
 
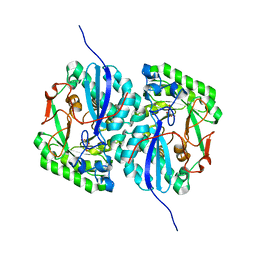 | | Crystal structure of L-rhamnonate dehydratase from Salmonella typhimurium complexed with Mg and D-malate | | Descriptor: | D-MALATE, MAGNESIUM ION, Putative galactonate dehydratase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-05-14 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of L-rhamnonate dehydratase from Salmonella typhimurium complexed with Mg and D-malate.
To be Published
|
|
4U0B
 
 | |
2QE7
 
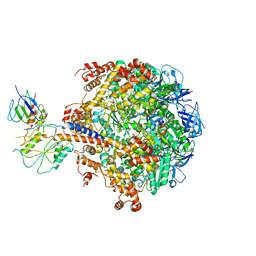 | | Crystal structure of the f1-atpase from the thermoalkaliphilic bacterium bacillus sp. ta2.a1 | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit epsilon, ... | | Authors: | Stocker, A, Keis, S, Vonck, J, Cook, G.M, Dimroth, P. | | Deposit date: | 2007-06-25 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | The Structural Basis for Unidirectional Rotation of Thermoalkaliphilic F(1)-ATPase.
Structure, 15, 2007
|
|
3D63
 
 | |
4JH0
 
 | | Crystal structure of dipeptidyl-peptidase 4 (CD26, adenosine deaminase complexing protein 2) (DPP-IV-WT) complex with bms-767778 AKA 2-(3-(aminomethyl)-4-(2,4- dichlorophenyl)-2-methyl-5-oxo-5,7-dihydro-6h-pyrrolo[3,4- b]pyridin-6-yl)-n,n-dimethylacetamide | | Descriptor: | 2-[3-(aminomethyl)-4-(2,4-dichlorophenyl)-2-methyl-5-oxo-5,7-dihydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-N,N-dimethylacetamide, Dipeptidyl peptidase 4 | | Authors: | Klei, H.E. | | Deposit date: | 2013-03-04 | | Release date: | 2013-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Optimization of Activity, Selectivity, and Liability Profiles in 5-Oxopyrrolopyridine DPP4 Inhibitors Leading to Clinical Candidate (Sa)-2-(3-(Aminomethyl)-4-(2,4-dichlorophenyl)-2-methyl-5-oxo-5H-pyrrolo[3,4-b]pyridin-6(7H)-yl)-N,N-dimethylacetamide (BMS-767778).
J.Med.Chem., 56, 2013
|
|
2GCT
 
 | |
4TVU
 
 | | Crystal structure of trehalose synthase from Deinococcus radiodurans reveals a closed conformation for catalysis of the intramolecular isomerization | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wang, Y.L, Chow, S.Y, Lin, Y.T, Liaw, S.H. | | Deposit date: | 2014-06-28 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of trehalose synthase from Deinococcus radiodurans reveal that a closed conformation is involved in catalysis of the intramolecular isomerization.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2GK1
 
 | | X-ray crystal structure of NGT-bound HexA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Beta-hexosaminidase subunit alpha, ... | | Authors: | Lemieux, M.J, Mark, B.L, Cherney, M.M, Withers, S.G, Mahuran, D.J, James, M.N. | | Deposit date: | 2006-03-31 | | Release date: | 2006-05-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystallographic Structure of Human beta-Hexosaminidase A: Interpretation of Tay-Sachs Mutations and Loss of G(M2) Ganglioside Hydrolysis.
J.Mol.Biol., 359, 2006
|
|
3EJB
 
 | |
4JD1
 
 | | Crystal Structure of Metallothiol Transferase FosB 2 from Bacillus anthracis str. Ames | | Descriptor: | FOSFOMYCIN, Metallothiol transferase FosB 2, TRIETHYLENE GLYCOL, ... | | Authors: | Maltseva, N, Kim, Y, Jedrzejczak, R, Sharma, S.V, Hamilton, C.J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-02-22 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Metallothiol Transferase FosB 2 from Bacillus anthracis str. Ames
To be Published
|
|
1PVE
 
 | |
3E4Q
 
 | | Crystal structure of apo DctB | | Descriptor: | C4-dicarboxylate transport sensor protein dctB, CALCIUM ION | | Authors: | Zhou, Y.F, Nan, J, Nan, B.Y, Liang, Y.H, Panjikar, S, Su, X.D. | | Deposit date: | 2008-08-12 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | C4-dicarboxylates sensing mechanism revealed by the crystal structures of DctB sensor domain.
J.Mol.Biol., 383, 2008
|
|
2GMU
 
 | | Crystal structure of E coli GDP-4-keto-6-deoxy-D-mannose-3-dehydratase complexed with PLP-glutamate ketimine intermediate | | Descriptor: | MAGNESIUM ION, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-D-GLUTAMIC ACID, Putative pyridoxamine 5-phosphate-dependent dehydrase, ... | | Authors: | Cook, P.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2006-04-07 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of GDP-4-keto-6-deoxy-D-mannose-3-dehydratase: a unique coenzyme B6-dependent enzyme.
Protein Sci., 15, 2006
|
|
4JFN
 
 | | Crystal structure of the N-terminal, growth factor-like domain of the amyloid precursor protein bound to copper | | Descriptor: | Amyloid beta A4 protein, COPPER (II) ION, GLYCEROL | | Authors: | Wild, K, Baumkotter, F, Kins, S. | | Deposit date: | 2013-02-28 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Amyloid precursor protein dimerization and synaptogenic function depend on copper binding to the growth factor-like domain
J.Neurosci., 34, 2014
|
|
4TXI
 
 | | Construct of MICAL-1 containing the monooxygenase and calponin homology domains | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Alqassim, S.S, Amzel, L.M, Bianchet, M.A. | | Deposit date: | 2014-07-03 | | Release date: | 2016-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.309 Å) | | Cite: | Modulation of MICAL Monooxygenase Activity by its Calponin Homology Domain: Structural and Mechanistic Insights.
Sci Rep, 6, 2016
|
|
2QHZ
 
 | | Crystal structure of protease inhibitor, MIT-1-AC87 in complex with wild type HIV-1 protease | | Descriptor: | (2E)-N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{(2-THIENYLMETHYL)[(2,4,5-TRIFLUOROPHENYL)SULFONYL]AMINO}PROPYL]-4,4,4-TRIFLUORO-3-METHYLBUT-2-ENAMIDE, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
3E70
 
 | | Structures and conformations in solution of the Signal Recognition Particle Receptor from the Archaeon Pyrococcus Furiosus | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Signal recognition particle receptor | | Authors: | Egea, P.F, Tsuruta, H, Napetschnig, J, Walter, P, Stroud, R.M. | | Deposit date: | 2008-08-17 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structures of the signal recognition particle receptor from the archaeon Pyrococcus furiosus: implications for the targeting step at the membrane.
Plos One, 3, 2008
|
|
2GK9
 
 | | Human Phosphatidylinositol-4-phosphate 5-kinase, type II, gamma | | Descriptor: | phosphatidylinositol-4-phosphate 5-kinase, type II, gamma | | Authors: | Uppenberg, J, Hogbom, M, Ogg, D, Arrowsmith, C, Berglund, H, Collins, R, Ehn, M, Flodin, S, Flores, A, Graslund, S, Holmberg-Schiavone, L, Edwards, A, Hammarstrom, M, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Hallberg, B.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-03-31 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Human Phosphatidylinositol-4-phosphate 5-kinase, type II, gamma
To be Published
|
|
3CF3
 
 | | Structure of P97/vcp in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Davies, J.M, Delabarre, B, Brunger, A.T, Weis, W.I. | | Deposit date: | 2008-03-01 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.25 Å) | | Cite: | Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change.
Structure, 16, 2008
|
|
2GQ3
 
 | | mycobacterium tuberculosis malate synthase in complex with magnesium, malate, and coenzyme A | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, COENZYME A, D-MALATE, ... | | Authors: | Anstrom, D.M, Remington, S.J. | | Deposit date: | 2006-04-19 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The product complex of M. tuberculosis malate synthase revisited.
Protein Sci., 15, 2006
|
|
2QHY
 
 | | Crystal Structure of protease inhibitor, MIT-1-AC86 in complex with wild type HIV-1 protease | | Descriptor: | ACETATE ION, N~2~-ACETYL-N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{(2-THIENYLMETHYL)[(2,4,5-TRIFLUOROPHENYL)SULFONYL]AMINO}PROPYL]-L-ALANINAMIDE, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
3CH9
 
 | |
