8TD8
 
 | | Structure of PYCR1 complexed with NADH and 2S-Hydroxy-3,3-dimethylbutyric acid | | Descriptor: | (2S)-2-hydroxy-3,3-dimethylbutanoic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
8TD5
 
 | | Structure of PYCR1 complexed with NADH and Tetrahydrothiophene-2-carboxylic acid | | Descriptor: | (2R)-thiolane-2-carboxylic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
5JXA
 
 | | Crystal structure of ligand-free VRC03 antigen-binding fragment. | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, PHOSPHATE ION, ... | | Authors: | Zhou, T, Moquin, S, Joyce, M.G, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-05-12 | | Release date: | 2016-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Somatic Hypermutation-Induced Changes in the Structure and Dynamics of HIV-1 Broadly Neutralizing Antibodies.
Structure, 24, 2016
|
|
6YU9
 
 | |
6O6E
 
 | | Crystal structure of PltF trapped with PltL using a proline adenosine vinylsulfonamide inhibitor | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-deoxy-5'-({(2S)-2-({2-[(N-{(2R)-4-[(dioxo-lambda~5~-phosphanyl)oxy]-2-hydroxy-3,3-dimethylbutanoyl}-beta-alanyl)amino]ethyl}sulfanyl)-2-[(2S)-pyrrolidin-2-yl]ethanesulfonyl}amino)adenosine, FORMIC ACID, ... | | Authors: | Corpuz, J.C, Podust, L.M. | | Deposit date: | 2019-03-06 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Dynamic visualization of type II peptidyl carrier protein recognition in pyoluteorin biosynthesis.
Rsc Chem Biol, 1, 2020
|
|
6HMC
 
 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 gene product) IN COMPLEX WITH THE INDENOINDOLE-TYPE INHIBITOR THN27 | | Descriptor: | 1,2-ETHANEDIOL, 5-propan-2-yl-4-prop-2-enoxy-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, Casein kinase II subunit alpha' | | Authors: | Niefind, K, Lindenblatt, D, Dimper, V, Jose, J, Le Borgne, M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Diacritic Binding of an Indenoindole Inhibitor by CK2 alpha Paralogs Explored by a Reliable Path to Atomic Resolution CK2 alpha ' Structures.
Acs Omega, 4, 2019
|
|
6NNO
 
 | |
8P7P
 
 | |
8P7O
 
 | |
5KNX
 
 | | Crystal structure of E. coli hypoxanthine phosphoribosyltransferase in complexed with {[(2-[(Hypoxanthin-9H-yl)methyl]propane-1,3-diyl)bis(oxy)]bis- (methylene)}diphosphonic Acid | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION, [2-[(6-oxidanylidene-1~{H}-purin-9-yl)methyl]-3-(phosphonomethoxy)propoxy]methylphosphonic acid | | Authors: | Eng, W.S, Keough, D.T, Hockova, D, Janeba, Z, Guddat, L.W. | | Deposit date: | 2016-06-28 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Acyclic Nucleoside Phosphonates in Complex with Escherichia coli Hypoxanthine Phosphoribosyltransferase
Chemistryselect, 1, 2016
|
|
8RPT
 
 | | Crystal structure of human DNPH1 mutant- D80A | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxynucleoside 5'-phosphate N-hydrolase 1, CHLORIDE ION, ... | | Authors: | Devi, S, da Silva, R.G. | | Deposit date: | 2024-01-17 | | Release date: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Human 2'-Deoxynucleoside 5'-Phosphate N-Hydrolase 1: The Catalytic Roles of Tyr24 and Asp80.
Chembiochem, 25, 2024
|
|
9INX
 
 | | Crystal structure of DAPK1 in complex with compound 10 | | Descriptor: | (~{E})-1-[2,4-bis(oxidanyl)phenyl]-3-(3-bromanyl-4-oxidanyl-phenyl)prop-2-en-1-one, Death-associated protein kinase 1, SULFATE ION | | Authors: | Yokoyama, T. | | Deposit date: | 2024-07-08 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery and optimization of isoliquiritigenin as a death-associated protein kinase 1 inhibitor.
Eur.J.Med.Chem., 279, 2024
|
|
9INW
 
 | | Crystal structure of DAPK1 in complex with compound 9 | | Descriptor: | (~{E})-1-[2,4-bis(oxidanyl)phenyl]-3-(3-chloranyl-4-oxidanyl-phenyl)prop-2-en-1-one, Death-associated protein kinase 1, SULFATE ION | | Authors: | Yokoyama, T. | | Deposit date: | 2024-07-08 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery and optimization of isoliquiritigenin as a death-associated protein kinase 1 inhibitor.
Eur.J.Med.Chem., 279, 2024
|
|
9U42
 
 | | Crystal Structure of Homogentisate 1,2-Dioxygenase from Acinetobacter in Complex with Zn ion | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glyoxalase, ZINC ION | | Authors: | Seo, P.-W, Hwangbo, S.-A, Park, S.-Y. | | Deposit date: | 2025-03-19 | | Release date: | 2025-07-23 | | Last modified: | 2025-11-19 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Mimicry Without Glyoxalase I Functional Convergence: A Homogentisate 1,2-Dioxygenase From Acinetobacter.
Proteins, 93, 2025
|
|
9RHW
 
 | | M. tuberculosis meets European Lead Factory: identification and structural characterization of novel Rv0183 inhibitors using X-ray crystallography: ELF1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Riegler-Berket, L, Goedl, L, Oberer, M, Sagmeister, T. | | Deposit date: | 2025-06-10 | | Release date: | 2025-08-13 | | Last modified: | 2025-10-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | M. tuberculosis meets European lead factory:Identification and structural characterization of novel Rv0183 inhibitors using X-ray crystallography
Disease and Therapeutics, 1, 2025
|
|
9NSZ
 
 | | OXA-23-NA-1-157, 6 minute soak | | Descriptor: | (5R)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase OXA-23, ... | | Authors: | Smith, C.A, Toth, M, Stewart, N.K, Vakulenko, S.B. | | Deposit date: | 2025-03-17 | | Release date: | 2025-08-06 | | Last modified: | 2025-10-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dual mechanism of the OXA-23 carbapenemase inhibition by the carbapenem NA-1-157.
Antimicrob.Agents Chemother., 69, 2025
|
|
8TYP
 
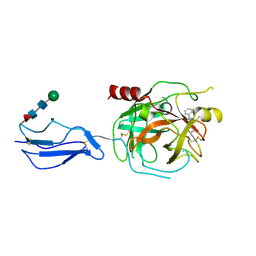 | | Complement Protease C1s Inhibited by 6-(4-phenylpiperazin-1-yl)pyridine-3-carboximidamide | | Descriptor: | 6-(4-phenylpiperazin-1-yl)pyridine-3-carboximidamide, Complement C1s subcomponent, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Geisbrecht, B.V. | | Deposit date: | 2023-08-25 | | Release date: | 2023-11-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of the C1s Protease and the Classical Complement Pathway by 6-(4-Phenylpiperazin-1-yl)Pyridine-3-Carboximidamide and Chemical Analogs.
J Immunol., 212, 2024
|
|
7PJN
 
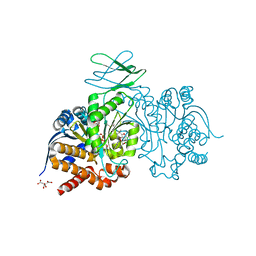 | | Crystal Structure of Ivosidenib-resistant IDH1 variant R132C S280F in complex with NADPH and inhibitor DS-1001B | | Descriptor: | (E)-3-(1-(5-(2-fluoropropan-2-yl)-3-(2,4,6-trichlorophenyl)isoxazole-4-carbonyl)-3-methyl-1H-indol-4-yl)acrylic acid, CITRIC ACID, GLYCEROL, ... | | Authors: | Reinbold, R, Rabe, P, Abboud, M.I, Schofield, C.J, Clifton, I.J. | | Deposit date: | 2021-08-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Resistance to the isocitrate dehydrogenase 1 mutant inhibitor ivosidenib can be overcome by alternative dimer-interface binding inhibitors.
Nat Commun, 13, 2022
|
|
6XSN
 
 | |
7SX7
 
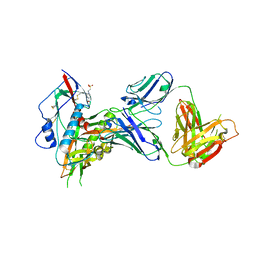 | |
6VNT
 
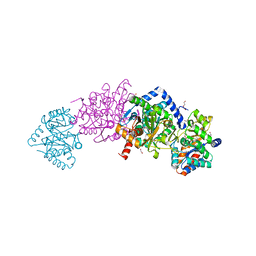 | | Tryptophan synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the alpha-site, aminoacrylate at the beta site, and sodium ion at the metal coordination site at 1.25 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, ... | | Authors: | Hilario, E, Fan, L, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Tryptophan synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the alpha-site, aminoacrylate at the beta site, and sodium ion at the metal coordination site at 1.25 Angstrom resolution.
To be Published
|
|
6YEH
 
 | | Arabidopsis thaliana glutamate dehydrogenase isoform 1 in apo form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Glutamate dehydrogenase 1, POTASSIUM ION | | Authors: | Ruszkowski, M, Grzechowiak, M, Jaskolski, M. | | Deposit date: | 2020-03-24 | | Release date: | 2020-05-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural Studies of Glutamate Dehydrogenase (Isoform 1) FromArabidopsis thaliana, an Important Enzyme at the Branch-Point Between Carbon and Nitrogen Metabolism.
Front Plant Sci, 11, 2020
|
|
5K8C
 
 | | X-ray structure of KdnB, 3-deoxy-alpha-D-manno-octulosonate 8-oxidase, from Shewanella oneidensis | | Descriptor: | 1,2-ETHANEDIOL, 3-deoxy-alpha-D-manno-octulosonate 8-oxidase, CHLORIDE ION, ... | | Authors: | Holden, H.M, Thoden, J.B, Zachman-Brockmeyer, T.R. | | Deposit date: | 2016-05-28 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of KdnB and KdnA from Shewanella oneidensis: Key Enzymes in the Formation of 8-Amino-3,8-Dideoxy-d-Manno-Octulosonic Acid.
Biochemistry, 55, 2016
|
|
8SBN
 
 | |
6HMD
 
 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 gene product) IN COMPLEX WITH THE INDENOINDOLE-TYPE INHIBITOR AR18 | | Descriptor: | 1,2-ETHANEDIOL, 5-[2-(diethylamino)ethyl]-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, CHLORIDE ION, ... | | Authors: | Niefind, K, Lindenblatt, D, Jose, J, Le Borgne, M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Diacritic Binding of an Indenoindole Inhibitor by CK2 alpha Paralogs Explored by a Reliable Path to Atomic Resolution CK2 alpha ' Structures.
Acs Omega, 4, 2019
|
|
