5AOP
 
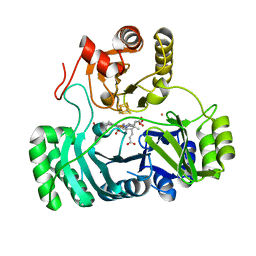 | | SULFITE REDUCTASE STRUCTURE REDUCED WITH CRII EDTA, 5-COORDINATE SIROHEME, SIROHEME FEII, [4FE-4S] +1 | | Descriptor: | IRON/SULFUR CLUSTER, POTASSIUM ION, SIROHEME, ... | | Authors: | Crane, B.R, Getzoff, E.D. | | Deposit date: | 1997-07-10 | | Release date: | 1998-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the siroheme- and Fe4S4-containing active center of sulfite reductase in different states of oxidation: heme activation via reduction-gated exogenous ligand exchange.
Biochemistry, 36, 1997
|
|
7GEP
 
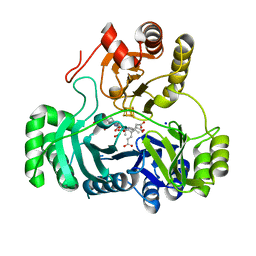 | | SULFITE REDUCTASE HEMOPROTEIN IN COMPLEX WITH A PARTIALLY OXIDIZED SULFIDE SPECIES | | Descriptor: | IRON/SULFUR CLUSTER, SIROHEME, SODIUM ION, ... | | Authors: | Crane, B.R, Getzoff, E.D. | | Deposit date: | 1997-07-11 | | Release date: | 1998-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the catalytic mechanism of sulfite reductase by X-ray crystallography: structures of the Escherichia coli hemoprotein in complex with substrates, inhibitors, intermediates, and products.
Biochemistry, 36, 1997
|
|
8VNZ
 
 | |
8VNV
 
 | |
6UZV
 
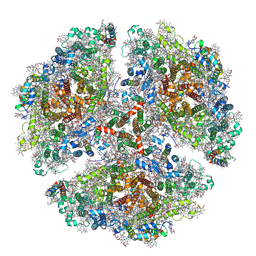 | | The structure of a red shifted photosystem I complex | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Toporik, H, Williams, D, Chiu, P.L, Mazor, Y. | | Deposit date: | 2019-11-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The structure of a red-shifted photosystem I reveals a red site in the core antenna.
Nat Commun, 11, 2020
|
|
6JW9
 
 | |
3GEO
 
 | | SULFITE REDUCTASE HEMOPROTEIN NITRITE COMPLEX | | Descriptor: | IRON/SULFUR CLUSTER, NITRITE ION, SIROHEME, ... | | Authors: | Crane, B.R, Getzoff, E.D. | | Deposit date: | 1997-07-10 | | Release date: | 1998-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the catalytic mechanism of sulfite reductase by X-ray crystallography: structures of the Escherichia coli hemoprotein in complex with substrates, inhibitors, intermediates, and products.
Biochemistry, 36, 1997
|
|
4GEP
 
 | | SULFITE REDUCTASE HEMOPROTEIN CYANIDE COMPLEX REDUCED WITH CRII EDTA | | Descriptor: | CYANIDE ION, IRON/SULFUR CLUSTER, POTASSIUM ION, ... | | Authors: | Crane, B.R, Getzoff, E.D. | | Deposit date: | 1997-07-10 | | Release date: | 1998-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the catalytic mechanism of sulfite reductase by X-ray crystallography: structures of the Escherichia coli hemoprotein in complex with substrates, inhibitors, intermediates, and products.
Biochemistry, 36, 1997
|
|
4AOP
 
 | |
1OAS
 
 | | O-ACETYLSERINE SULFHYDRYLASE FROM SALMONELLA TYPHIMURIUM | | Descriptor: | O-ACETYLSERINE SULFHYDRYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Burkhard, P, Rao, G.S.J, Hohenester, E, Schnackerz, K.D, Cook, P.F, Jansonius, J.N. | | Deposit date: | 1999-01-29 | | Release date: | 2000-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of O-acetylserine sulfhydrylase from Salmonella typhimurium.
J.Mol.Biol., 283, 1998
|
|
3AOP
 
 | | SULFITE REDUCTASE HEMOPROTEIN PHOTOREDUCED WITH PROFLAVINE EDTA, SIROHEME FEII,[4FE-4S] +1, PHOSPHATE BOUND | | Descriptor: | IRON/SULFUR CLUSTER, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Crane, B.R, Getzoff, E.D. | | Deposit date: | 1997-07-09 | | Release date: | 1998-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the siroheme- and Fe4S4-containing active center of sulfite reductase in different states of oxidation: heme activation via reduction-gated exogenous ligand exchange.
Biochemistry, 36, 1997
|
|
3RR2
 
 | |
6NTE
 
 | |
4GIG
 
 | |
2AOP
 
 | | SULFITE REDUCTASE: REDUCED WITH CRII EDTA, SIROHEME FEII, [4FE-4S] +1, PHOSPHATE BOUND | | Descriptor: | IRON/SULFUR CLUSTER, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Crane, B.R, Getzoff, E.D. | | Deposit date: | 1997-07-09 | | Release date: | 1998-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of the siroheme- and Fe4S4-containing active center of sulfite reductase in different states of oxidation: heme activation via reduction-gated exogenous ligand exchange.
Biochemistry, 36, 1997
|
|
2KND
 
 | | Psb27 structure from Synechocystis | | Descriptor: | Photosystem II 11 kDa protein | | Authors: | Cormann, K.U, Nowaczyk, M.M, Bangert, J.-A, Ikeuchi, M, Roegner, M, Stoll, R. | | Deposit date: | 2009-08-21 | | Release date: | 2009-09-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of Psb27 in solution: implications for transient binding to photosystem II during biogenesis and repair
Biochemistry, 48, 2009
|
|
2E03
 
 | |
4GUM
 
 | | Cystal structure of locked-trimer of human MIF | | Descriptor: | CHLORIDE ION, Macrophage migration inhibitory factor | | Authors: | Fan, C, Lolis, E. | | Deposit date: | 2012-08-29 | | Release date: | 2013-07-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | MIF intersubunit disulfide mutant antagonist supports activation of CD74 by endogenous MIF trimer at physiologic concentrations.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3OXL
 
 | | Human lysine methyltransferase Smyd3 in complex with AdoHcy (Form II) | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SET and MYND domain-containing protein 3, ZINC ION | | Authors: | Xu, S, Wu, J, Sun, B, Zhong, C, Ding, J. | | Deposit date: | 2010-09-21 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural and biochemical studies of human lysine methyltransferase Smyd3 reveal the important functional roles of its post-SET and TPR domains and the regulation of its activity by DNA binding
Nucleic Acids Res., 39, 2011
|
|
3OXF
 
 | | Human lysine methyltransferase Smyd3 in complex with AdoHcy (Form I) | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SET and MYND domain-containing protein 3, ZINC ION | | Authors: | Xu, S, Wu, J, Sun, B, Zhong, C, Ding, J. | | Deposit date: | 2010-09-21 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural and biochemical studies of human lysine methyltransferase Smyd3 reveal the important functional roles of its post-SET and TPR domains and the regulation of its activity by DNA binding.
Nucleic Acids Res., 39, 2011
|
|
3QWV
 
 | | Crystal structure of histone lysine methyltransferase SmyD2 in complex with the cofactor product AdoHcy | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SET and MYND domain-containing protein 2, ZINC ION | | Authors: | Jiang, Y, Sirinupong, N, Brunzelle, J, Yang, Z. | | Deposit date: | 2011-02-28 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structures of histone and p53 methyltransferase SmyD2 reveal a conformational flexibility of the autoinhibitory C-terminal domain.
Plos One, 6, 2011
|
|
3OXG
 
 | | human lysine methyltransferase Smyd3 in complex with AdoHcy (Form III) | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SET and MYND domain-containing protein 3, ZINC ION | | Authors: | Xu, S, Wu, J, Sun, B, Zhong, C, Ding, J. | | Deposit date: | 2010-09-21 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Structural and biochemical studies of human lysine methyltransferase Smyd3 reveal the important functional roles of its post-SET and TPR domains and the regulation of its activity by DNA binding.
Nucleic Acids Res., 39, 2011
|
|
1QNT
 
 | |
2GEP
 
 | | SULFITE REDUCTASE HEMOPROTEIN, OXIDIZED, SIROHEME FEIII [4FE-4S] +2,SULFITE COMPLEX | | Descriptor: | IRON/SULFUR CLUSTER, SIROHEME, SODIUM ION, ... | | Authors: | Crane, B.R, Getzoff, E.D. | | Deposit date: | 1997-07-10 | | Release date: | 1998-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the catalytic mechanism of sulfite reductase by X-ray crystallography: structures of the Escherichia coli hemoprotein in complex with substrates, inhibitors, intermediates, and products.
Biochemistry, 36, 1997
|
|
5AUK
 
 | | Crystal structure of the Ga-substituted Ferredoxin | | Descriptor: | BENZAMIDINE, Ferredoxin-1, SULFATE ION, ... | | Authors: | Kurisu, G, Muraki, N, Taya, M, Hase, T. | | Deposit date: | 2015-04-24 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | X-ray Structure and Nuclear Magnetic Resonance Analysis of the Interaction Sites of the Ga-Substituted Cyanobacterial Ferredoxin
Biochemistry, 54, 2015
|
|
