7OVY
 
 | | Crystal structure of a dimeric based inhibitor JG34 in complex with the MMP-12 catalytic domain | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Macrophage metalloelastase, ... | | Authors: | Ciccone, L, Rossello, A, Vera, L, Nuti, E, Stura, E.A. | | Deposit date: | 2021-06-15 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Discovery of Dimeric Arylsulfonamides as Potent ADAM8 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
8BL1
 
 | | Human Sirt6 in complex with the inhibitor S6039 and ADP-ribose | | Descriptor: | 2-azanyl-5-[4-[[5-(2-fluorophenyl)thiophen-2-yl]methyl]piperazin-1-yl]sulfonyl-benzenesulfonamide, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | You, W, Steegborn, C. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Development of novel Sirtuin 6 inhibitors and activators based on a protein crystallography-based fragment screen
To Be Published
|
|
6WNM
 
 | | The structure of Pf4r from a superinfective isolate of the filamentous phage Pf4 of Pseudomonas aeruginosa PA01 | | Descriptor: | Pf4r, [(2R)-3-{[2-(carboxymethoxy)benzene-1-carbonyl]amino}-2-methoxypropyl](hydroxy)mercury | | Authors: | Michie, K.A, Norrian, P, Duggin, I.G, McDougald, D, Rice, S.A. | | Deposit date: | 2020-04-22 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The Repressor C Protein, Pf4r, Controls Superinfection of Pseudomonas aeruginosa PAO1 by the Pf4 Filamentous Phage and Regulates Host Gene Expression.
Viruses, 13, 2021
|
|
6X0P
 
 | | Ash1L SET domain Q2265A mutant in complex with AS-5 | | Descriptor: | 3-[6-(aminomethyl)-1-(2-hydroxyethyl)-1H-indol-3-yl]benzene-1-carbothioamide, Histone-lysine N-methyltransferase ASH1L, S-ADENOSYLMETHIONINE, ... | | Authors: | Rogawski, D.S, Li, H, Borkin, D, Cierpicki, T, Grembecka, J. | | Deposit date: | 2020-05-17 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of first-in-class inhibitors of ASH1L histone methyltransferase with anti-leukemic activity.
Nat Commun, 12, 2021
|
|
6L36
 
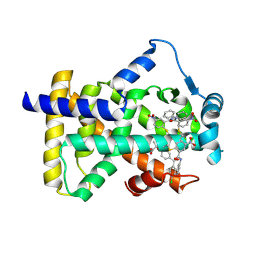 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-fenofibric acid co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, 2-chloro-5-nitro-N-phenylbenzamide, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-10-09 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6WQ1
 
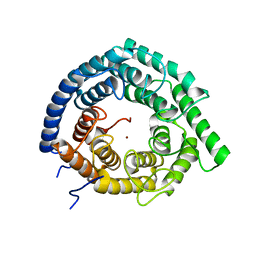 | | Eukaryotic LanCL2 protein | | Descriptor: | LanC-like protein 2, ZINC ION | | Authors: | Nair, S.K, Garg, N. | | Deposit date: | 2020-04-28 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | LanCLs add glutathione to dehydroamino acids generated at phosphorylated sites in the proteome.
Cell, 184, 2021
|
|
6LD4
 
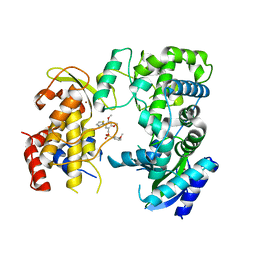 | | Zika NS5 polymerase domain | | Descriptor: | 2,4-dimethoxy-5-thiophen-2-yl-benzoic acid, 3-methoxybenzenesulfonamide, RNA-directed RNA polymerase NS5, ... | | Authors: | El Sahili, A, Lescar, J. | | Deposit date: | 2019-11-20 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Non-nucleoside Inhibitors of Zika Virus RNA-Dependent RNA Polymerase.
J.Virol., 94, 2020
|
|
6E4F
 
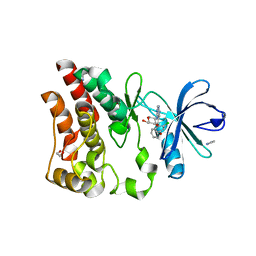 | | Crystal structure of ARQ 531 in complex with the kinase domain of BTK | | Descriptor: | 1,2-ETHANEDIOL, 1,5-anhydro-2-{[5-(2-chloro-4-phenoxybenzene-1-carbonyl)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]amino}-2,3,4-trideoxy-D-erythro-hexitol, IMIDAZOLE, ... | | Authors: | Eathiraj, S. | | Deposit date: | 2018-07-17 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The BTK Inhibitor ARQ 531 Targets Ibrutinib-Resistant CLL and Richter Transformation.
Cancer Discov, 8, 2018
|
|
6X21
 
 | | Crystal structure of PT1673 bound to HIF2a-B*:ARNT-B* complex | | Descriptor: | 1-(3-bromo-5-fluorophenoxy)-4-[(difluoromethyl)sulfonyl]-2-nitrobenzene, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Du, X. | | Deposit date: | 2020-05-19 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of PT1673 bound to HIF2a-B*:ARNT-B* complex
To Be Published
|
|
1EO7
 
 | | BACILLUS CIRCULANS STRAIN 251 CYCLODEXTRIN GLYCOSYLTRANSFERASE IN COMPLEX WITH MALTOHEXAOSE | | Descriptor: | CALCIUM ION, PROTEIN (CYCLODEXTRIN GLYCOSYLTRANSFERASE), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Uitdehaag, J.C.M, Dijkstra, B.W. | | Deposit date: | 2000-03-22 | | Release date: | 2000-11-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structures of maltohexaose and maltoheptaose bound at the donor sites of cyclodextrin glycosyltransferase give insight into the mechanisms of transglycosylation activity and cyclodextrin size specificity.
Biochemistry, 39, 2000
|
|
4Q02
 
 | | Second-site screening of K-Ras in the presence of covalently attached first-site ligands | | Descriptor: | 3,4-difluorobenzenethiol, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Sun, Q, Phan, J, Friberg, A, Camper, D.V, Olejniczak, E.T, Fesik, S.W. | | Deposit date: | 2014-03-31 | | Release date: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | A method for the second-site screening of K-Ras in the presence of a covalently attached first-site ligand.
J.Biomol.Nmr, 60, 2014
|
|
1J3F
 
 | | Crystal Structure of an Artificial Metalloprotein:Cr(III)(3,3'-Me2-salophen)/apo-A71G Myoglobin | | Descriptor: | 'N,N'-BIS-(2-HYDROXY-3-METHYL-BENZYLIDENE)-BENZENE-1,2-DIAMINE', CHROMIUM ION, Myoglobin, ... | | Authors: | Koshiyama, T, Kono, M, Ohashi, M, Ueno, T, Suzuki, A, Yamane, T, Watanabe, Y. | | Deposit date: | 2003-01-24 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Coordinated Design of Cofactor and Active Site Structures in Development of New Protein Catalysts
J.Am.Chem.Soc., 127, 2005
|
|
6ELP
 
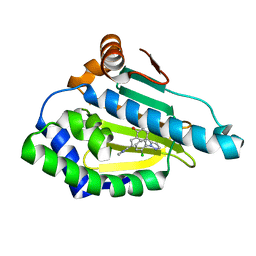 | |
4WPF
 
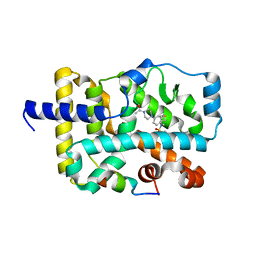 | | Crystal structure of RORc in complex with a phenyl sulfonamide agonist | | Descriptor: | N-[4-(4-acetylpiperazin-1-yl)-2-fluorobenzyl]-N-cyclobutylbenzenesulfonamide, Nuclear receptor ROR-gamma, RHKILHRLLQEGSPS | | Authors: | Kiefer, J.R, Wallweber, H.A, de Leon Boenig, G, Hymowitz, S.G. | | Deposit date: | 2014-10-18 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Minor Structural Change to Tertiary Sulfonamide RORc Ligands Led to Opposite Mechanisms of Action.
Acs Med.Chem.Lett., 6, 2015
|
|
6E97
 
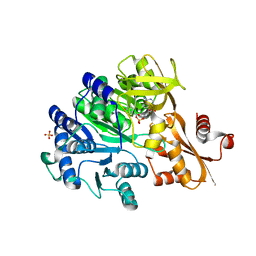 | |
4Q13
 
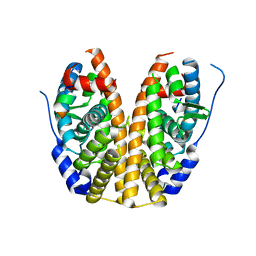 | |
4WHR
 
 | | Anhydride reaction intermediate trapped in Protocatechuate 3,4-dioxygenase (pseudomonas putida) at pH 8.5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-fluorobenzene-1,2-diol, 4-fluorooxepine-2,7-dione, ... | | Authors: | Knoot, C.J, Lipscomb, J.D. | | Deposit date: | 2014-09-23 | | Release date: | 2014-12-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structures of alkylperoxo and anhydride intermediates in an intradiol ring-cleaving dioxygenase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6LOX
 
 | | Crystal Structure of human glutaminase with macrocyclic inhibitor | | Descriptor: | (E)-15,22-Dioxa-4,11-diaza-5(2,5)-thiadiazola-10(3,6)-pyridazina-1,14(1,3)-dibenzenacyclodocosaphan-18-ene-3,12-dione, Glutaminase kidney isoform, mitochondrial | | Authors: | Bian, J, Li, Z, Xu, X, Wang, J, Li, L. | | Deposit date: | 2020-01-07 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-Enabled Discovery of Novel Macrocyclic Inhibitors Targeting Glutaminase 1 Allosteric Binding Site.
J.Med.Chem., 64, 2021
|
|
6EHR
 
 | |
5ZS8
 
 | |
6LLK
 
 | | Biphenyl-2,2',3-triol-soaked terminal oxygenase of carbazole 1,9a-dioxygenase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-(2-hydroxyphenyl)benzene-1,2-diol, DIMETHYL SULFOXIDE, ... | | Authors: | Wang, Y.X, Suzuki-Minakuchi, C, Nojiri, H. | | Deposit date: | 2019-12-23 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biphenyl-2,2',3-triol-soaked terminal oxygenase of carbazole 1,9a-dioxygenase
To Be Published
|
|
4PXM
 
 | |
1B8Y
 
 | | X-RAY STRUCTURE OF HUMAN STROMELYSIN CATALYTIC DOMAIN COMPLEXED WITH NON-PEPTIDE INHIBITORS: IMPLICATIONS FOR INHIBITOR SELECTIVITY | | Descriptor: | CALCIUM ION, PROTEIN (STROMELYSIN-1), SULFATE ION, ... | | Authors: | Pavlovsky, A.G, Williams, M.G, Ye, Q.-Z, Ortwine, D.F, Purchase II, C.F, White, A.D, Dhanaraj, V, Roth, B.D, Johnson, L.L, Hupe, D, Humblet, C, Blundell, T.L. | | Deposit date: | 1999-02-03 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of human stromelysin catalytic domain complexed with nonpeptide inhibitors: implications for inhibitor selectivity.
Protein Sci., 8, 1999
|
|
4PWJ
 
 | | Crystal structure of V30M mutant human transthyretin complexed with nordihydroguaiaretic acid | | Descriptor: | 4-[(2R,3S)-3-[(3,4-DIHYDROXYPHENYL)METHYL]-2-METHYLBUTYL]BENZENE-1,2-DIOL, Transthyretin | | Authors: | Yokoyama, T, Kosaka, Y, Mizuguchi, M. | | Deposit date: | 2014-03-20 | | Release date: | 2014-11-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Inhibitory Activities of Propolis and Its Promising Component, Caffeic Acid Phenethyl Ester, against Amyloidogenesis of Human Transthyretin
J.Med.Chem., 57, 2014
|
|
6LX4
 
 | | X-ray structure of human PPARalpha ligand binding domain-fenofibric acid co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
