7DX3
 
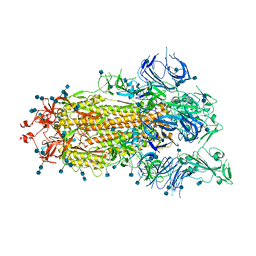 | | S protein of SARS-CoV-2 bound with PD of ACE2 in the conformation 1 (1 up RBD and no PD bound) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
7DX1
 
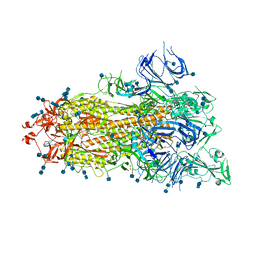 | | S protein of SARS-CoV-2 D614G mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
7DX0
 
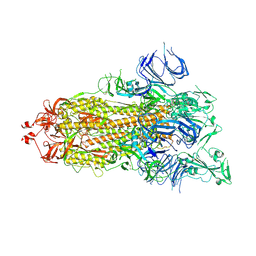 | | Trypsin-digested S protein of SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
7DWZ
 
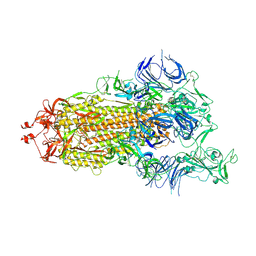 | | S protein of SARS-CoV-2 in the active conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
5DWS
 
 | | Crystal Structure of ITCH WW3 domain in complex with TXNIP peptide | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog, UNKNOWN ATOM OR ION, txnip | | Authors: | Liu, Y, Tempel, W, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-22 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of ITCH WW3 domain in complex with TXNIP peptide
to be published
|
|
7DX2
 
 | | Trypsin-digested S protein of SARS-CoV-2 D614G mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
7DX8
 
 | | Trypsin-digested S protein of SARS-CoV-2 bound with PD of ACE2 in the conformation 2 (2 up RBD and 2 PD bound) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
9B38
 
 | | Kainate receptor GluK2 in complex with agonist glutamate with pseudo 4-fold symmetrical ligand-binding domain layer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Nadezhdin, K.D, Gangwar, S.P, Sobolevsky, A.I. | | Deposit date: | 2024-03-18 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Kainate receptor channel opening and gating mechanism.
Nature, 630, 2024
|
|
7DVQ
 
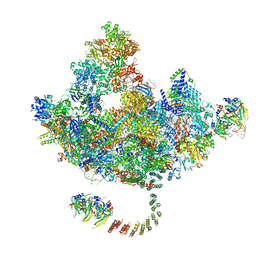 | | Cryo-EM Structure of the Activated Human Minor Spliceosome (minor Bact Complex) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'-O-[(S)-hydroxy{[(R)-hydroxy{[(S)-hydroxy(methoxy)phosphoryl]oxy}phosphoryl]oxy}phosphoryl]guanosine, Armadillo repeat-containing protein 7, ... | | Authors: | Bai, R, Wan, R, Wang, L, Xu, K, Zhang, Q, Lei, J, Shi, Y. | | Deposit date: | 2021-01-14 | | Release date: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of the activated human minor spliceosome.
Science, 371, 2021
|
|
7DX5
 
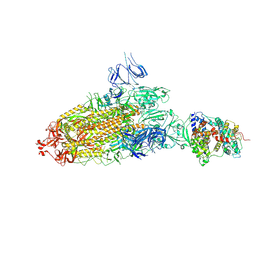 | | S protein of SARS-CoV-2 bound with PD of ACE2 in the conformation 2 (1 up RBD and 1 PD bound) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
7DX9
 
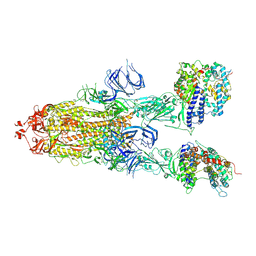 | | Trypsin-digested S protein of SARS-CoV-2 bound with PD of ACE2 in the conformation 3 (3 up RBD and 2 PD bound) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
9B36
 
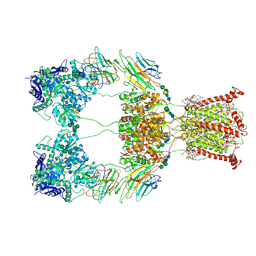 | | Open state of kainate receptor GluK2 in complex with agonist glutamate and positive allosteric modulator BPAM344 bound to two concanavalin A dimers. Composite map. | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nadezhdin, K.D, Gangwar, S.P, Sobolevsky, A.I. | | Deposit date: | 2024-03-18 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.29 Å) | | Cite: | Kainate receptor channel opening and gating mechanism.
Nature, 630, 2024
|
|
7DX7
 
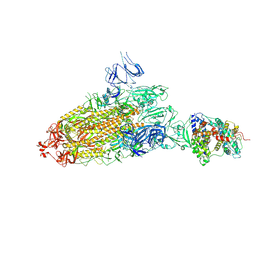 | | Trypsin-digested S protein of SARS-CoV-2 bound with PD of ACE2 in the conformation 1 (1 up RBD and 1 PD bound) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
4UJC
 
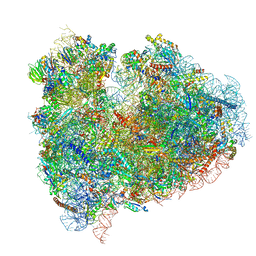 | | mammalian 80S HCV-IRES initiation complex with eIF5B POST-like state | | Descriptor: | 18S RIBOSOMAL RNA, 28S RIBOSOMAL RNA, 40S RIBOSOMAL PROTEIN ES1, ... | | Authors: | Yamamoto, H, Unbehaun, A, Loerke, J, Behrmann, E, Marianne, C, Burger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2014-06-18 | | Release date: | 2014-07-30 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Structure of the Mammalian 80S Initiation Complex with Initiation Factor 5B on Hcv-Ires RNA.
Nat.Struct.Mol.Biol., 21, 2014
|
|
5DOX
 
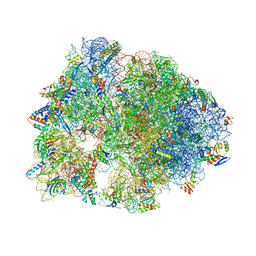 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with Hygromycin-A at 3.1A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Polikanov, Y.S, Starosta, A.L, Juette, M.F, Altman, R.B, Terry, D.S, Lu, W, Burnett, B.J, Dinos, G, Reynolds, K, Blanchard, S.C, Steitz, T.A, Wilson, D.N. | | Deposit date: | 2015-09-11 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Distinct tRNA Accommodation Intermediates Observed on the Ribosome with the Antibiotics Hygromycin A and A201A.
Mol.Cell, 58, 2015
|
|
5E1B
 
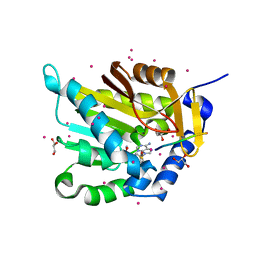 | | Crystal structure of NRMT1 in complex with SPKRIA peptide | | Descriptor: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-29 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
9AVA
 
 | | Co-crystal structure of human TREX1 in complex with an inhibitor | | Descriptor: | (2R)-2-[(5R,6S,8R,9aS)-8-amino-1-oxo-5-(2-phenylethyl)hexahydro-1H-pyrrolo[1,2-a][1,4]diazepin-2(3H)-yl]-N-[(3,4-dichlorophenyl)methyl]-4-methylpentanamide, POTASSIUM ION, Three-prime repair exonuclease 1, ... | | Authors: | Dehghani-Tafti, S, Dong, A, Li, Y, Xu, J, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-01 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Co-crystal structure of human TREX1 in complex with an inhibitor
To be published
|
|
4UQ8
 
 | | Electron cryo-microscopy of bovine Complex I | | Descriptor: | ACYL CARRIER PROTEIN, MITOCHONDRIAL, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Vinothkumar, K.R, Zhu, J, Hirst, J. | | Deposit date: | 2014-06-21 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.95 Å) | | Cite: | Architecture of Mammalian Respiratory Complex I.
Nature, 515, 2014
|
|
6NFN
 
 | | Fab fragment of anti-cocaine antibody h2E2 bound to benzoylecgonine | | Descriptor: | 3-(BENZOYLOXY)-8-METHYL-8-AZABICYCLO[3.2.1]OCTANE-2-CARBOXYLIC ACID, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pokkuluri, P.R, Tan, K. | | Deposit date: | 2018-12-20 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural analysis of free and liganded forms of the Fab fragment of a high-affinity anti-cocaine antibody, h2E2.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
4UJ3
 
 | | Crystal structure of human Rab11-Rabin8-FIP3 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, RAB-3A-INTERACTING PROTEIN, ... | | Authors: | Vetter, M, Lorentzen, E. | | Deposit date: | 2015-04-08 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Rab11-Fip3-Rabin8 Reveals Simultaneous Binding of Fip3 and Rabin8 Effectors to Rab11.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5E57
 
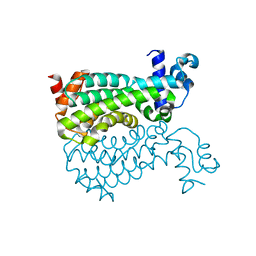 | | Crystal structure of Mycobacterium smegmatis AmtR | | Descriptor: | ACETATE ION, Transcription regulator AmtR | | Authors: | Vickers, C.J, McKenzie, J.L, Arcus, V.L. | | Deposit date: | 2015-10-07 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure and Function of AmtR in Mycobacterium smegmatis: Implications for Post-Transcriptional Regulation of Urea Metabolism through a Small Antisense RNA.
J.Mol.Biol., 428, 2016
|
|
4UM8
 
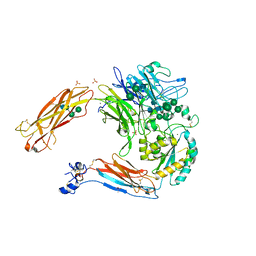 | | Crystal structure of alpha V beta 6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, ... | | Authors: | Dong, X, Springer, T.A. | | Deposit date: | 2014-05-15 | | Release date: | 2014-11-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.852 Å) | | Cite: | Structural Determinants of Integrin Beta-Subunit Specificity for Latent Tgf-Beta
Nat.Struct.Mol.Biol., 21, 2014
|
|
6NH8
 
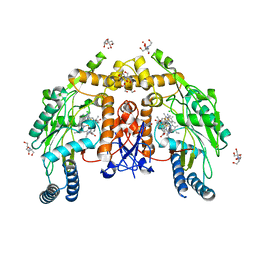 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 6-(5-(3-(dimethylamino)propyl)-2,3,4-trifluorophenethyl)-4-methylpyridin-2-amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{5-[3-(dimethylamino)propyl]-2,3,4-trifluorophenyl}ethyl)-4-methylpyridin-2-amine, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2018-12-21 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Optimization of Blood-Brain Barrier Permeability with Potent and Selective Human Neuronal Nitric Oxide Synthase Inhibitors Having a 2-Aminopyridine Scaffold.
J. Med. Chem., 62, 2019
|
|
9BJU
 
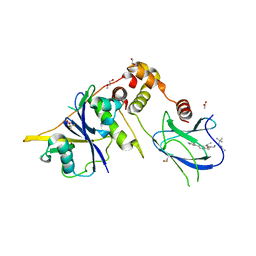 | | Crystal structure of the complex between VHL, ElonginB, ElonginC, and compound 5 | | Descriptor: | 1,2-ETHANEDIOL, 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, ACETATE ION, ... | | Authors: | Murray, J.M, Wu, H, Fuhrmann, J, Fairbrother, W.J, DiPasquale, A. | | Deposit date: | 2024-04-25 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Potency-enhanced peptidomimetic VHL ligands with improved oral bioavailability
To Be Published
|
|
7E80
 
 | | Cryo-EM structure of the flagellar rod with hook and export apparatus from Salmonella | | Descriptor: | Flagellar MS ring L1, Flagellar MS ring L2, Flagellar basal body rod protein FlgB, ... | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2021-02-28 | | Release date: | 2021-04-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
