5M9O
 
 | | Crystal structure of human SND1 extended Tudor domain in complex with a symmetrically dimethylated E2F peptide | | Descriptor: | E2F peptide, Staphylococcal nuclease domain-containing protein 1 | | Authors: | Tallant, C, Savitsky, P, Moehlenbrink, J, Chan, C, Nunez-Alonso, G, Siejka, P, Sorrell, F.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, La Thangue, N.B, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-11-01 | | Release date: | 2016-12-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of human SND1 extended Tudor domain in complex with a symmetrically
dimethylated E2F peptide
To Be Published
|
|
5MDS
 
 | |
6TMQ
 
 | | Rapid optimisation of fragments and hits to lead compounds from screening of crude reaction mixtures | | Descriptor: | DIMETHYL SULFOXIDE, N-(2-AMINOETHYL)-2-{3-CHLORO-4-[(4-ISOPROPYLBENZYL)OXY]PHENYL} ACETAMIDE, [2,4-bis(oxidanyl)phenyl]-[(1~{S})-6,7-dimethoxy-1-pyridin-3-yl-3,4-dihydro-1~{H}-isoquinolin-2-yl]methanone, ... | | Authors: | Baker, L.M, Aimon, A, Murray, J.B, Surgenor, A.E, Matassova, N, Roughley, S.D, von Delft, F, Hubbard, R.E. | | Deposit date: | 2019-12-05 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | Rapid optimisation of fragments and hits to lead compounds from screening of crude reaction mixtures
Commun Chem, 2020
|
|
6TJ5
 
 | | T. gondii myosin A trimeric complex with ELC1 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Calmodulin, ... | | Authors: | Pazicky, S, Loew, C. | | Deposit date: | 2019-11-25 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Structural role of essential light chains in the apicomplexan glideosome.
Commun Biol, 3, 2020
|
|
6TJA
 
 | | Crystal structure of the SVS_A2 protein (W79F,G83L mutant) from ancestral sequence reconstruction at 2.27 A resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, SVS_variant_AS1 | | Authors: | Rudraraju, R, Schnell, R, Schneider, G. | | Deposit date: | 2019-11-25 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Engineering of Ancestors as a Tool to Elucidate Structure, Mechanism, and Specificity of Extant Terpene Cyclase.
J.Am.Chem.Soc., 143, 2021
|
|
6TJH
 
 | | Crystal structure of the computationally designed Cake9 protein | | Descriptor: | Cake9, GLYCEROL, SULFATE ION | | Authors: | Mylemans, B, Laier, I, Noguchi, H, Voet, A.R.D. | | Deposit date: | 2019-11-26 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Febs J., 288, 2021
|
|
5MEQ
 
 | | Human Leukocyte Antigen A02 presenting ILAKFLHTL | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Lloyd, A, Crowther, M, Sewell, A.K. | | Deposit date: | 2016-11-16 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural Mechanism Underpinning Cross-reactivity of a CD8+ T-cell Clone That Recognizes a Peptide Derived from Human Telomerase Reverse Transcriptase.
J. Biol. Chem., 292, 2017
|
|
6TOZ
 
 | | Crystal structure of Bacillus paralicheniformis alpha-amylase in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ACETIC ACID, Amylase, ... | | Authors: | Rozeboom, H.J, Janssen, D.B. | | Deposit date: | 2019-12-12 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Characterization of the starch surface binding site on Bacillus paralicheniformis alpha-amylase.
Int.J.Biol.Macromol., 165, 2020
|
|
6TPR
 
 | | PqsR (MvfR) bound to inhibitory compound 40 | | Descriptor: | 2-[(5-methyl-[1,2,4]triazino[5,6-b]indol-3-yl)sulfanyl]-~{N}-(4-pyridin-2-yloxyphenyl)ethanamide, Transcriptional regulator MvfR | | Authors: | Richardson, W.K, Emsley, J. | | Deposit date: | 2019-12-14 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Hit Identification of New Potent PqsR Antagonists as Inhibitors of Quorum Sensing in Planktonic and Biofilm GrownPseudomonas aeruginosa.
Front Chem, 8, 2020
|
|
5MH5
 
 | | D-2-hydroxyacid dehydrogenases (D2-HDH) from Haloferax mediterranei in complex with 2-keto-hexanoic acid and NADP+ (1.4 A resolution) | | Descriptor: | 2-Ketohexanoic acid, D-2-hydroxyacid dehydrogenase, MAGNESIUM ION, ... | | Authors: | Bisson, C, Baker, P.J, Domenech Perez, J, Pramanpol, N, Harding, S.E, Rice, D.W, Ferrer, J. | | Deposit date: | 2016-11-23 | | Release date: | 2018-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Productive ternary complexes of D-2-hydroxyacid dehydrogenase provide insights into the chiral specificity of its reaction mechanism
To Be Published
|
|
5MHQ
 
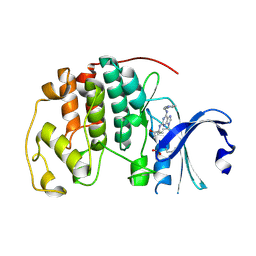 | | CCT068127 in complex with CDK2 | | Descriptor: | (2~{R},3~{S})-3-[[9-propan-2-yl-6-(pyridin-3-ylmethylamino)purin-2-yl]amino]pentan-2-ol, Cyclin-dependent kinase 2 | | Authors: | Whittaker, S.R, Barlow, C, Martin, M.P, Mancusi, C, Wagner, S, Barrie, E, te Poele, R, Sharp, S, Brown, N, Wilson, S, Clarke, P, Walton, M.I, MacDonald, E, Blagg, J, Noble, M.E.M, Garrett, M.D, Workman, P. | | Deposit date: | 2016-11-25 | | Release date: | 2017-12-20 | | Last modified: | 2018-03-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular profiling and combinatorial activity of CCT068127: a potent CDK2 and CDK9 inhibitor.
Mol Oncol, 12, 2018
|
|
6TQS
 
 | | The crystal structure of the MSP domain of human MOSPD2 in complex with the conventional FFAT motif of ORP1. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Di Mattia, T, Wendling, C, Cavarelli, J, Tomasetto, C, Alpy, F. | | Deposit date: | 2019-12-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts.
Embo J., 39, 2020
|
|
6T1U
 
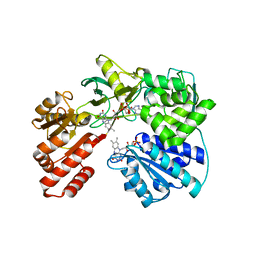 | | Cytochrome P450 reductase from Candida tropicalis | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADPH--cytochrome P450 reductase | | Authors: | Opperman, D.J, Sewell, B.T. | | Deposit date: | 2019-10-06 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biochemical and structural insights into the cytochrome P450 reductase from Candida tropicalis.
Sci Rep, 9, 2019
|
|
5MI7
 
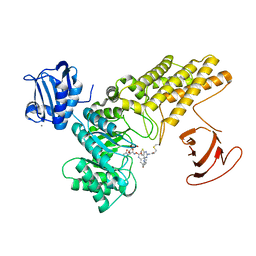 | | BtGH84 mutant with covalent modification by MA4 in complex with PUGNAc | | Descriptor: | CALCIUM ION, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE, O-GlcNAcase BT_4395, ... | | Authors: | Darby, J.F, Davies, G.J, Hubbard, R.E. | | Deposit date: | 2016-11-27 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Increase of enzyme activity through specific covalent modification with fragments.
Chem Sci, 8, 2017
|
|
6T2N
 
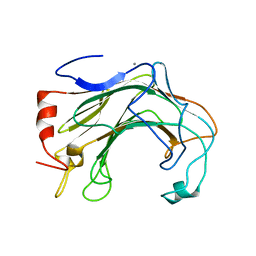 | | Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown | | Descriptor: | CALCIUM ION, Glycoside hydrolase family 16 protein | | Authors: | Crouch, L.I, Liberato, M.V, Ubranowicz, P.A, Basle, A, Lamb, C.A, Cooke, K, Doona, M, Needham, S, Brady, R.R, Berrington, J.E, Madubic, K, Chater, P, Zhang, F, Linhardt, R.J, Spence, D.I.R, Bolam, D.N. | | Deposit date: | 2019-10-09 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown.
Nat Commun, 11, 2020
|
|
6T2X
 
 | |
5MKT
 
 | | Crystal structure of mouse prorenin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin-1 | | Authors: | Yan, Y, Read, R. | | Deposit date: | 2016-12-05 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of mouse prorenin
To Be Published
|
|
5MMF
 
 | | Crystal Structure of CK2alpha with Compound 7 bound | | Descriptor: | (3-chloranyl-4-phenyl-phenyl)methyl-propyl-azanium, ACETATE ION, Casein kinase II subunit alpha | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2016-12-09 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066.
Bioorg. Med. Chem., 25, 2017
|
|
6TS3
 
 | |
5MMW
 
 | | Crystal structure of the Retinoid X Receptor alpha in complex with synthetic honokiol derivative 6 and a fragment of the TIF2 co-activator. | | Descriptor: | (~{E})-3-[3-(2-methyl-3-phenyl-phenyl)-4-oxidanyl-phenyl]prop-2-enoic acid, LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN, Retinoic acid receptor RXR-alpha | | Authors: | Andrei, S.A, Brunsveld, L, Scheepstra, M, Ottmann, C. | | Deposit date: | 2016-12-12 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ligand Dependent Switch from RXR Homo- to RXR-NURR1 Heterodimerization.
ACS Chem Neurosci, 8, 2017
|
|
6TS9
 
 | | Crystal structure of GES-5 carbapenemase | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Beta-lactamase, ... | | Authors: | Maso, L, Tondi, D, Klein, R, Montanari, M, Bellio, C, Celenza, G, Brenk, R, Cendron, L. | | Deposit date: | 2019-12-20 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Targeting the Class A Carbapenemase GES-5 via Virtual Screening.
Biomolecules, 10, 2020
|
|
5MLB
 
 | | Crystal structure of human RAS in complex with darpin K27 | | Descriptor: | DARPin K27, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Debreczeni, J.E, Guillard, S, Kolasinska-Zwierz, P, Breed, J, Zhang, J, Bery, N, Marwood, R, Tart, J, Overman, R, Stocki, P, Mistry, B, Phillips, C, Rabbitts, T, Jackson, R, Minter, R. | | Deposit date: | 2016-12-06 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | INhibition of RAS nucleotide exchange by a DARPin: structural characterisation and effects on downstream signalling by active RAS
To Be Published
|
|
6T69
 
 | | Crystal structure of Toxoplasma gondii Morn1(V shape) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Membrane occupation and recognition nexus protein MORN1, ... | | Authors: | Grishkovskaya, I, Kostan, J, Sajko, S, Morriswood, B, Djinovic-Carugo, K. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats.
Plos One, 15, 2020
|
|
6T36
 
 | | Crystal structure of the PTPN3 PDZ domain bound to the HBV core protein C-terminal peptide | | Descriptor: | BROMIDE ION, Capsid protein, Tyrosine-protein phosphatase non-receptor type 3 | | Authors: | Genera, M, Mechaly, A, Haouz, A, Caillet-Saguy, C. | | Deposit date: | 2019-10-10 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Molecular basis of the interaction of the human tyrosine phosphatase PTPN3 with the hepatitis B virus core protein.
Sci Rep, 11, 2021
|
|
6T6B
 
 | | Crystal structure of PPARgamma in complex with compound 16 (MF27) | | Descriptor: | (2~{R})-2-[[6-[(2,4-dichlorophenyl)sulfonylamino]-1,3-benzothiazol-2-yl]sulfanyl]octanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Ni, X, Hanke, T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-18 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Selective Modulator of Peroxisome Proliferator-Activated Receptor gamma with an Unprecedented Binding Mode.
J.Med.Chem., 63, 2020
|
|
