6SZH
 
 | | Acinetobacter baumannii undecaprenyl pyrophosphate synthase (AB-UppS) in complex with GW197 | | Descriptor: | 3,5-dimethyl-1~{H}-pyrrole-2-carbonitrile, CALCIUM ION, Ditrans,polycis-undecaprenyl-diphosphate synthase ((2E,6E)-farnesyl-diphosphate specific) | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-10-02 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Cocktailed fragment screening by X-ray crystallography of the antibacterial target undecaprenyl pyrophosphate synthase from Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6SPW
 
 | | Structure of protein kinase CK2 catalytic subunit with the CK2beta-competitive bisubstrate inhibitor ARC3140 | | Descriptor: | 8-[4,5,6,7-tetrakis(iodanyl)benzimidazol-1-yl]octanoic acid, ARC3140, Casein kinase II subunit alpha, ... | | Authors: | Niefind, K, Schnitzler, A. | | Deposit date: | 2019-09-03 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Unexpected CK2 beta-antagonistic functionality of bisubstrate inhibitors targeting protein kinase CK2.
Bioorg.Chem., 96, 2020
|
|
6SZW
 
 | |
6SQ7
 
 | | Crystal structure of M. tuberculosis InhA in complex with NAD+ and 2-(4-chloro-3-nitrobenzoyl)benzoic acid | | Descriptor: | 2-(4-chloranyl-3-nitro-phenyl)carbonylbenzoic acid, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mendes, V, Sabbah, M, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Fragment-Based Design ofMycobacterium tuberculosisInhA Inhibitors.
J.Med.Chem., 63, 2020
|
|
6T0Y
 
 | |
6SG4
 
 | | Structure of CDK2/cyclin A M246Q, S247EN | | Descriptor: | Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Salamina, M, Basle, A, Massa, B, Noble, M.E.M, Endicott, J.A. | | Deposit date: | 2019-08-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discriminative SKP2 Interactions with CDK-Cyclin Complexes Support a Cyclin A-Specific Role in p27KIP1 Degradation.
J.Mol.Biol., 433, 2021
|
|
6SQY
 
 | | Mouse dCTPase in complex with dCMP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, MAGNESIUM ION, dCTP pyrophosphatase 1 | | Authors: | Scaletti, E.R, Claesson, M, Helleday, H, Jemth, A.S, Stenmark, P. | | Deposit date: | 2019-09-04 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The First Structure of an Active Mammalian dCTPase and its Complexes With Substrate Analogs and Products.
J.Mol.Biol., 432, 2020
|
|
6SHH
 
 | |
6SRD
 
 | |
5OP4
 
 | | Structure of CHK1 10-pt. mutant complex with aminopyrimidine LRRK2 inhibitor | | Descriptor: | Serine/threonine-protein kinase Chk1, [4-[[4-(ethylamino)-5-(trifluoromethyl)pyrimidin-2-yl]amino]-2-fluoranyl-5-methoxy-phenyl]-morpholin-4-yl-methanone | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-09 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
5OPD
 
 | |
5OPW
 
 | | Crystal structure of the GroEL mutant A109C | | Descriptor: | 60 kDa chaperonin | | Authors: | Yan, X, Shi, Q, Bracher, A, Milicic, G, Singh, A.K, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2017-08-10 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | GroEL Ring Separation and Exchange in the Chaperonin Reaction.
Cell, 172, 2018
|
|
6SI1
 
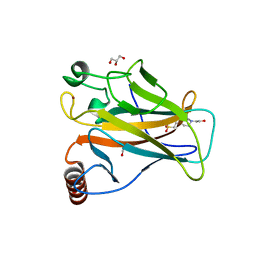 | | p53 cancer mutant Y220H | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cellular tumor antigen p53, ... | | Authors: | Joerger, A.C, Kraemer, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-08 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Targeting Cavity-Creating p53 Cancer Mutations with Small-Molecule Stabilizers: the Y220X Paradigm.
Acs Chem.Biol., 15, 2020
|
|
5OR9
 
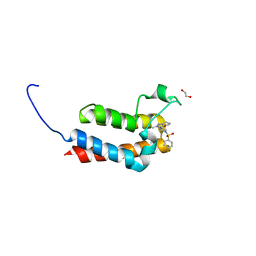 | | Crystal Structure of BAZ2B bromodomain in complex with 1-methyl-cyclopentapyrazole compound 13 | | Descriptor: | (2~{S})-1-(4-fluorophenyl)sulfonyl-~{N}-(2-methyl-5,6-dihydro-4~{H}-cyclopenta[c]pyrazol-3-yl)pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Lolli, G, Vedove, A.D, Marchand, J.-R, Caflisch, A. | | Deposit date: | 2017-08-15 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Inhibitors of Four Bromodomains by Fragment-Anchored Ligand Docking.
J Chem Inf Model, 57, 2017
|
|
6SIY
 
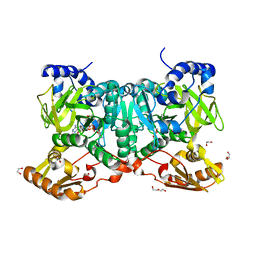 | | PaaK family AMP-ligase with AMP and substrate | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXYANTHRANILIC ACID, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5ONF
 
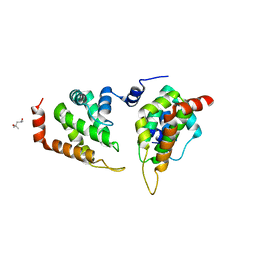 | | The ENTH domain from epsin-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Epsin-1 | | Authors: | Garcia-Alai, M, GIeras, A, Meijers, R. | | Deposit date: | 2017-08-03 | | Release date: | 2018-03-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Epsin and Sla2 form assemblies through phospholipid interfaces.
Nat Commun, 9, 2018
|
|
5ONK
 
 | | Native YndL | | Descriptor: | CITRATE ANION, IMIDAZOLE, SULFATE ION, ... | | Authors: | Ramaswamy, S, Rasheed, M, Morelli, C, Calvio, C, Sutton, B, Pastore, A. | | Deposit date: | 2017-08-03 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | The structure of PghL hydrolase bound to its substrate poly-gamma-glutamate.
FEBS J., 285, 2018
|
|
6SL2
 
 | | ALPHA-ACTININ FROM ENTAMOEBA HISTOLYTICA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Calponin homology domain protein putative, ... | | Authors: | Pinotsis, N, Khan, M.B, Djinovic-Carugo, K. | | Deposit date: | 2019-08-18 | | Release date: | 2020-08-26 | | Last modified: | 2020-11-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Calcium modulates the domain flexibility and function of an alpha-actinin similar to the ancestral alpha-actinin.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5ONS
 
 | | Crystal structure of the minimal DENR-MCTS1 complex | | Descriptor: | Density-regulated protein, GLYCEROL, Malignant T-cell-amplified sequence 1, ... | | Authors: | Ahmed, Y.L, Sinning, I. | | Deposit date: | 2017-08-04 | | Release date: | 2018-05-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | DENR-MCTS1 heterodimerization and tRNA recruitment are required for translation reinitiation.
PLoS Biol., 16, 2018
|
|
5ONY
 
 | | As-isolated resting state copper nitrite reductase from Achromobacter xylosoxidans | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Halsted, T.P, Yamashita, K, Hirata, K, Ago, H, Ueno, G, Tosha, T, Eady, R.R, Antonyuk, S.V, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2017-08-04 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An unprecedented dioxygen species revealed by serial femtosecond rotation crystallography in copper nitrite reductase.
IUCrJ, 5, 2018
|
|
6SLJ
 
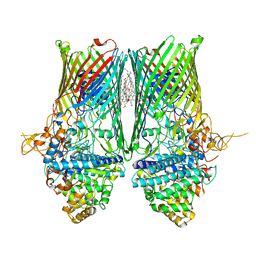 | | Structure of the RagAB peptide transporter | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ALA-SER-THR-THR-GLY-ALA-ASN-SER-GLN-ARG, ... | | Authors: | Madej, M, Ranson, N.A, White, J.B.R. | | Deposit date: | 2019-08-20 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structural and functional insights into oligopeptide acquisition by the RagAB transporter from Porphyromonas gingivalis.
Nat Microbiol, 5, 2020
|
|
5OO5
 
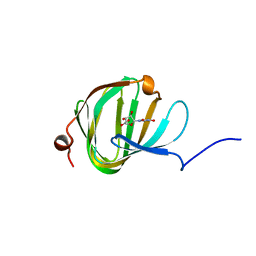 | | Streptomyces PAC13 with uridine uronic acid | | Descriptor: | Putative cupin_2 domain-containing isomerase, uridine uronic acid | | Authors: | Chung, C, Michailidou, F. | | Deposit date: | 2017-08-05 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Pac13 is a Small, Monomeric Dehydratase that Mediates the Formation of the 3'-Deoxy Nucleoside of Pacidamycins.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6SND
 
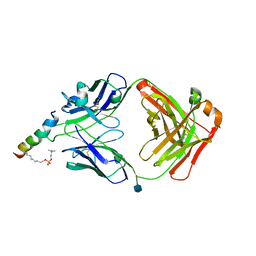 | | crystal structure of LN01 Fab in complex with an HIV-1 gp41 peptide | | Descriptor: | 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Caillat, C, Pinto, D, Corti, D, Fenwick, C, Pantaleo, G, Weissenhorn, W. | | Deposit date: | 2019-08-23 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for Broad HIV-1 Neutralization by the MPER-Specific Human Broadly Neutralizing Antibody LN01.
Cell Host Microbe, 26, 2019
|
|
5MLG
 
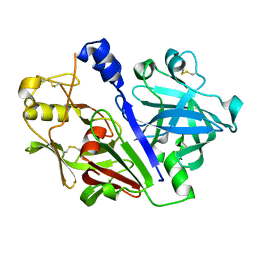 | | Crystal structure of rat prorenin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin | | Authors: | Yan, Y, Read, R. | | Deposit date: | 2016-12-06 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of rat prorenin
To Be Published
|
|
6T3G
 
 | |
