2KIZ
 
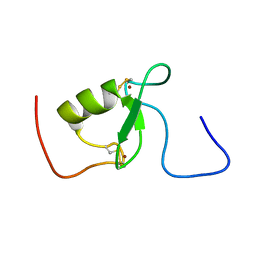 | | Solution structure of Arkadia RING-H2 finger domain | | Descriptor: | E3 ubiquitin-protein ligase Arkadia, ZINC ION | | Authors: | Kandias, N.G, Chasapis, C.T, Bentrop, D, Episkopou, V, Spyroulias, G.A. | | Deposit date: | 2009-05-13 | | Release date: | 2010-05-19 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NMR-based insights into the conformational and interaction properties of Arkadia RING-H2 E3 Ub ligase.
Proteins, 80, 2012
|
|
2FCW
 
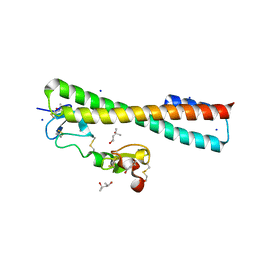 | | Structure of a Complex Between the Pair of the LDL Receptor Ligand-Binding Modules 3-4 and the Receptor Associated Protein (RAP). | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha-2-macroglobulin receptor-associated protein, CALCIUM ION, ... | | Authors: | Beglova, N, Fisher, C, Blacklow, S.C. | | Deposit date: | 2005-12-12 | | Release date: | 2006-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structure of an LDLR-RAP Complex Reveals a General Mode for Ligand Recognition by Lipoprotein Receptors
Mol.Cell, 22, 2006
|
|
6BKG
 
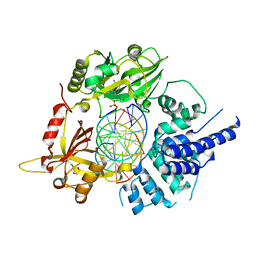 | | Human LigIV catalytic domain with bound DNA-adenylate intermediate in closed conformation | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Moon, A.F, Tumbale, P.P, Schellenberg, M.J, Williams, R.S, Williams, J.G, Kunkel, T.A, Pedersen, L.C, Bebenek, B. | | Deposit date: | 2017-11-08 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Structures of DNA-bound human ligase IV catalytic core reveal insights into substrate binding and catalysis.
Nat Commun, 9, 2018
|
|
3X2I
 
 | | X-ray structure of PcCel45A N92D apo form at 298K. | | Descriptor: | 3-methylpentane-1,5-diol, Endoglucanase V-like protein | | Authors: | Nakamura, A, Ishida, T, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
6BKF
 
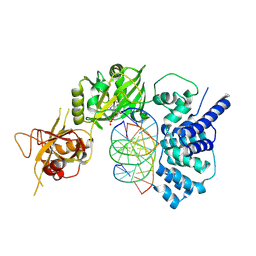 | | Lysyl-adenylate form of human LigIV catalytic domain with bound DNA substrate in open conformation | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*C)-3'), DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*GP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Moon, A.F, Tumbale, P.P, Schellenberg, M.J, Williams, R.S, Williams, J.G, Kunkel, T.A, Pedersen, L.C, Bebenek, B. | | Deposit date: | 2017-11-08 | | Release date: | 2018-07-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structures of DNA-bound human ligase IV catalytic core reveal insights into substrate binding and catalysis.
Nat Commun, 9, 2018
|
|
2AWH
 
 | | Human Nuclear Receptor-Ligand Complex 1 | | Descriptor: | Peroxisome proliferator activated receptor delta, VACCENIC ACID, heptyl beta-D-glucopyranoside | | Authors: | Fyffe, S.A, Alphey, M.S, Buetow, L, Smith, T.K, Ferguson, M.A.J, Sorensen, M.D, Bjorkling, F, Hunter, W.N. | | Deposit date: | 2005-09-01 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recombinant Human PPAR-beta/delta Ligand-binding Domain is Locked in an Activated Conformation by Endogenous Fatty Acids
J.Mol.Biol., 356, 2006
|
|
2B50
 
 | | Human Nuclear Receptor-Ligand Complex 2 | | Descriptor: | CALCIUM ION, Peroxisome proliferator activated receptor delta, VACCENIC ACID, ... | | Authors: | Fyffe, S.A, Alphey, M.S, Buetow, L, Smith, T.K, Ferguson, M.A.J, Sorensen, M.D, Bjorkling, F, Hunter, W.N. | | Deposit date: | 2005-09-27 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recombinant Human PPAR-beta/delta Ligand-binding Domain is Locked in an Activated Conformation by Endogenous Fatty Acids
J.Mol.Biol., 356, 2006
|
|
2LGV
 
 | | Rbx1 | | Descriptor: | E3 ubiquitin-protein ligase RBX1, ZINC ION | | Authors: | Spratt, D.E, Shaw, G.S. | | Deposit date: | 2011-08-02 | | Release date: | 2012-03-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Selective recruitment of an e2~ubiquitin complex by an e3 ubiquitin ligase.
J.Biol.Chem., 287, 2012
|
|
252L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, T4 LYSOZYME | | Authors: | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | Deposit date: | 1997-10-28 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
1LDK
 
 | | Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF Ubiquitin Ligase Complex | | Descriptor: | CULLIN HOMOLOG, CYCLIN A/CDK2-ASSOCIATED PROTEIN P19, SKP2-like protein type gamma, ... | | Authors: | Zheng, N, Schulman, B.A, Song, L, Miller, J.J, Jeffrey, P.D, Wang, P, Chu, C, Koepp, D.M, Elledge, S.J, Pagano, M, Conaway, R.C, Conaway, J.W, Harper, J.W, Pavletich, N.P. | | Deposit date: | 2002-04-08 | | Release date: | 2002-05-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex.
Nature, 416, 2002
|
|
8DP2
 
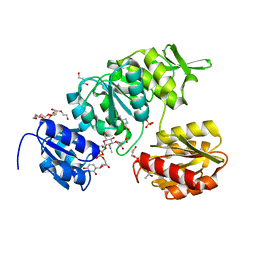 | | Crystal Structure of UDP-N-acetylmuramoylalanine--D-glutamate ligase (MurD) from Pseudomonas aeruginosa PAO1 in complex with UMA (Uridine-5'-diphosphate-N-acetylmuramoyl-L-Alanine) | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-07-14 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of UDP-N-acetylmuramoylalanine--D-glutamate ligase (MurD) from Pseudomonas aeruginosa PAO1 in complex with UMA (Uridine-5'-diphosphate-N-acetylmuramoyl-L-Alanine)
to be published
|
|
1UPW
 
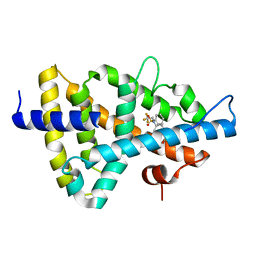 | | Crystal structure of the human Liver X receptor beta ligand binding domain in complex with a synthetic agonist | | Descriptor: | N-(2,2,2-TRIFLUOROETHYL)-N-{4-[2,2,2-TRIFLUORO-1-HYDROXY-1-(TRIFLUOROMETHYL)ETHYL]PHENYL}BENZENESULFONAMIDE, OXYSTEROLS RECEPTOR LXR-BETA | | Authors: | Hoerer, S, Schmid, A, Heckel, A, Budzinski, R.M, Nar, H. | | Deposit date: | 2003-10-13 | | Release date: | 2004-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Human Liver X Receptor Beta Ligand-Binding Domain in Complex with a Synthetic Agonist
J.Mol.Biol., 334, 2003
|
|
7SIR
 
 | |
5HEH
 
 | |
1EVT
 
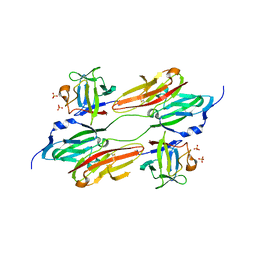 | | CRYSTAL STRUCTURE OF FGF1 IN COMPLEX WITH THE EXTRACELLULAR LIGAND BINDING DOMAIN OF FGF RECEPTOR 1 (FGFR1) | | Descriptor: | PROTEIN (FIBROBLAST GROWTH FACTOR 1), PROTEIN (FIBROBLAST GROWTH FACTOR RECEPTOR 1), SULFATE ION | | Authors: | Plotnikov, A.N, Hubbard, S.R, Schlessinger, J, Mohammadi, M. | | Deposit date: | 2000-04-20 | | Release date: | 2000-05-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of two FGF-FGFR complexes reveal the determinants of ligand-receptor specificity.
Cell(Cambridge,Mass.), 101, 2000
|
|
6DO3
 
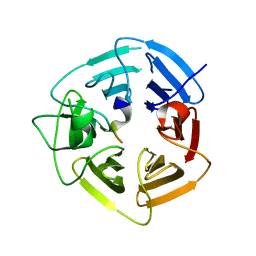 | | KLHDC2 ubiquitin ligase in complex with SelK C-end degron | | Descriptor: | Kelch domain-containing protein 2, SelK C-end Degron | | Authors: | Rusnac, D.V, Lin, H.C, Yen, H.C.S, Zheng, N. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.165 Å) | | Cite: | Recognition of the Diglycine C-End Degron by CRL2KLHDC2Ubiquitin Ligase.
Mol. Cell, 72, 2018
|
|
5VZW
 
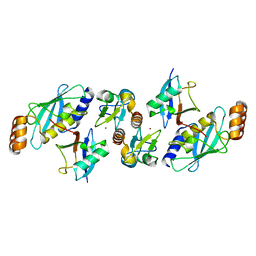 | | TRIM23 RING domain in complex with UbcH5-Ub | | Descriptor: | E3 ubiquitin-protein ligase TRIM23, Ubiquitin, Ubiquitin-conjugating enzyme E2 D2, ... | | Authors: | Pornillos, O, Dawidziak, D. | | Deposit date: | 2017-05-29 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.278 Å) | | Cite: | Structure and catalytic activation of the TRIM23 RING E3 ubiquitin ligase.
Proteins, 85, 2017
|
|
3FS0
 
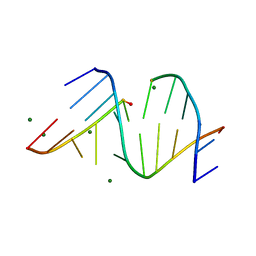 | |
6A5W
 
 | | FXR-LBD with HNC143 and SRC1 | | Descriptor: | 2-[2-[[3-[2,6-bis(chloranyl)phenyl]-5-cyclopropyl-1,2-oxazol-4-yl]methoxy]-6-azaspiro[3.4]octan-6-yl]-1,3-benzothiazole-6-carboxylic acid, Bile acid receptor, Nuclear receptor coactivator 1 | | Authors: | Wang, N, Liu, J. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Ligand binding and heterodimerization with retinoid X receptor alpha (RXR alpha ) induce farnesoid X receptor (FXR) conformational changes affecting coactivator binding
J. Biol. Chem., 293, 2018
|
|
3FTM
 
 | |
1YCB
 
 | | DISTAL POCKET POLARITY IN LIGAND BINDING TO MYOGLOBIN: DEOXY AND CARBONMONOXY FORMS OF A THREONINE68 (E11) MUTANT INVESTIGATED BY X-RAY CRYSTALLOGRAPHY AND INFRARED SPECTROSCOPY | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Cameron, A.D, Smerdon, S.J, Wilkinson, A.J, Habash, J, Helliwell, J.R. | | Deposit date: | 1993-08-10 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distal pocket polarity in ligand binding to myoglobin: deoxy and carbonmonoxy forms of a threonine68(E11) mutant investigated by X-ray crystallography and infrared spectroscopy.
Biochemistry, 32, 1993
|
|
6A5Y
 
 | | Crystal structure of human FXR/RXR-LBD heterodimer bound to HNC143 and 9cRA and SRC1 | | Descriptor: | (9cis)-retinoic acid, 2-[2-[[3-[2,6-bis(chloranyl)phenyl]-5-cyclopropyl-1,2-oxazol-4-yl]methoxy]-6-azaspiro[3.4]octan-6-yl]-1,3-benzothiazole-6-carboxylic acid, Bile acid receptor, ... | | Authors: | Wang, N, Liu, J. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand binding and heterodimerization with retinoid X receptor alpha (RXR alpha ) induce farnesoid X receptor (FXR) conformational changes affecting coactivator binding
J. Biol. Chem., 293, 2018
|
|
6VE8
 
 | | Structure of the Glutamate-Like Receptor GLR3.2 ligand-binding domain in complex with Methionine | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gangwar, S.P, Green, M.N, Yoder, J.B, Sobolevsky, A.I. | | Deposit date: | 2019-12-30 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Arabidopsis Glutamate Receptor-like Channel GLR3.2 Ligand-Binding Domain.
Structure, 29, 2021
|
|
6VEA
 
 | | Structure of the Glutamate-Like Receptor GLR3.2 ligand-binding domain in complex with Glycine | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCINE, Glutamate receptor 3.2, ... | | Authors: | Gangwar, S.P, Green, M.N, Yoder, J.B, Sobolevsky, A.I. | | Deposit date: | 2019-12-30 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure of the Arabidopsis Glutamate Receptor-like Channel GLR3.2 Ligand-Binding Domain.
Structure, 29, 2021
|
|
6A60
 
 | | Crystal structure of human FXR/RXR-LBD heterodimer bound to GW4064 and 9cRA and SRC1 | | Descriptor: | (9cis)-retinoic acid, 3-[(E)-2-(2-chloro-4-{[3-(2,6-dichlorophenyl)-5-(1-methylethyl)isoxazol-4-yl]methoxy}phenyl)ethenyl]benzoic acid, Bile acid receptor, ... | | Authors: | Wang, N, Liu, J. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Ligand binding and heterodimerization with retinoid X receptor alpha (RXR alpha ) induce farnesoid X receptor (FXR) conformational changes affecting coactivator binding
J. Biol. Chem., 293, 2018
|
|
