5N2W
 
 | | WT-Parkin and pUB complex | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin,E3 ubiquitin-protein ligase parkin, Polyubiquitin-B, ... | | Authors: | Kumar, A, Chaugule, V.K, Johnson, C, Toth, R, Sundaramoorthy, R, Knebel, A, Walden, H. | | Deposit date: | 2017-02-08 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Parkin-phosphoubiquitin complex reveals cryptic ubiquitin-binding site required for RBR ligase activity.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5N5E
 
 | |
5NHE
 
 | |
5N87
 
 | | TTK kinase domain in complex with NTRC 0066-0 | | Descriptor: | 2,5,8,11-TETRAOXATRIDECANE, Dual specificity protein kinase TTK, SODIUM ION, ... | | Authors: | Uitdehaag, J, Willemsen-Seegers, N, Sterrenburg, J.G, de Man, J, Buijsman, R.C, Zaman, G.J.R. | | Deposit date: | 2017-02-23 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Target Residence Time-Guided Optimization on TTK Kinase Results in Inhibitors with Potent Anti-Proliferative Activity.
J. Mol. Biol., 429, 2017
|
|
5N8Q
 
 | | Structure of truncated Norcoclaurine Synthase from Thalictrum flavum | | Descriptor: | S-norcoclaurine synthase | | Authors: | Sula, A, Lichman, B.R, Pesnot, T, Ward, J.M, Hailes, H.C, Keep, N.H. | | Deposit date: | 2017-02-24 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Evidence for the Dopamine-First Mechanism of Norcoclaurine Synthase.
Biochemistry, 56, 2017
|
|
6RVQ
 
 | | SaFtsz-GDP-EthGLy | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cell division protein FtsZ, ... | | Authors: | Fernandez-Tornero, C, Andreu, J.M. | | Deposit date: | 2019-05-31 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.136 Å) | | Cite: | Nucleotide-induced folding of cell division protein FtsZ from Staphylococcus aureus.
Febs J., 287, 2020
|
|
5NJC
 
 | | E. coli Microcin-processing metalloprotease TldD/E (TldD E263A mutant) with hexapeptide bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Metalloprotease PmbA, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
5N90
 
 | | Crystal Structure of Drosophila DHX36 helicase in complex with TTGTGGTGT | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*TP*TP*GP*TP*GP*GP*TP*GP*T)-3'), ... | | Authors: | Chen, W.-F, Rety, S, Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.069 Å) | | Cite: | Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Structure, 26, 2018
|
|
5N9L
 
 | | Crystal structure of human Protein kinase CK2 catalytic subunit in complex with the ATP-competitive dibenzofuran inhibitor TF (4b) | | Descriptor: | (4~{Z})-6,7-bis(chloranyl)-4-[[(4-methylphenyl)amino]methylidene]-8-oxidanyl-1,2-dihydrodibenzofuran-3-one, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Schnitzler, A, Gratz, A, Bollacke, A, Weyrich, M, Kucklaender, U, Wuensch, B, Goetz, C, Niefind, K, Jose, J. | | Deposit date: | 2017-02-25 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A pi-Halogen Bond of Dibenzofuranones with the Gatekeeper Phe113 in Human Protein Kinase CK2 Leads to Potent Tight Binding Inhibitors.
Pharmaceuticals, 11, 2018
|
|
6RJS
 
 | | Inter-dimeric interface controls function and stability of S-methionine adenosyltransferase from U. urealiticum | | Descriptor: | Methionine adenosyltransferase | | Authors: | Shahar, A, Zarivach, R, Bershtein, S, Kleiner, D, Shmulevich, F. | | Deposit date: | 2019-04-29 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The interdimeric interface controls function and stability of Ureaplasma urealiticum methionine S-adenosyltransferase.
J.Mol.Biol., 431, 2019
|
|
5NK5
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 1m | | Descriptor: | 2-[[3-[(3-aminophenyl)carbamoyl]phenyl]amino]-~{N}-(2-chloranyl-6-methyl-phenyl)-1,3-thiazole-5-carboxamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.329 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5NKD
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 2i | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3~{R})-1-[3-[[5-[(2-chloranyl-6-methyl-phenyl)carbamoyl]-1,3-thiazol-2-yl]amino]phenyl]carbonylpyrrolidin-3-yl]ethanoic acid, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.408 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
6RLH
 
 | |
6RLQ
 
 | | CRYSTAL STRUCTURE OF THE HUMAN PRMT5:MEP50 COMPLEX with JNJ45031882 | | Descriptor: | (1~{S},2~{R},3~{S},5~{R})-3-[2-(2-azanyl-3-bromanyl-quinolin-7-yl)ethyl]-5-(4-azanylpyrrolo[2,3-d]pyrimidin-7-yl)cyclop entane-1,2-diol, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | Brown, D.G, Robinson, C.M, Pande, V. | | Deposit date: | 2019-05-02 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | CRYSTAL STRUCTURE OF THE HUMAN PRMT5:MEP50 COMPLEX with JNJ45031882
To Be Published
|
|
6RZF
 
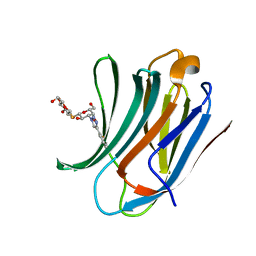 | | Galectin-3C in complex with ortho-fluoroaryltriazole galactopyranosyl 1-thio-D-glucopyranoside derivative | | Descriptor: | (2~{S},3~{R},4~{S},5~{S},6~{R})-2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(2-fluorophenyl)-1,2,3-triazol-1-yl]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]sulfanyl-6-(hydroxymethyl)oxane-3,4,5-triol, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.016 Å) | | Cite: | Entropy-Entropy Compensation between the Protein, Ligand, and Solvent Degrees of Freedom Fine-Tunes Affinity in Ligand Binding to Galectin-3C.
Jacs Au, 1, 2021
|
|
6RZK
 
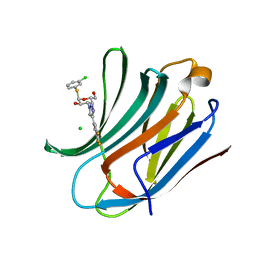 | | Galectin-3C in complex with trifluoroaryltriazole monothiogalactoside derivative:3(Chlorine) | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{R})-2-(3-chlorophenyl)sulfanyl-6-(hydroxymethyl)-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, R, Verteramo, M.L, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.046 Å) | | Cite: | Thermodynamic studies of halogen-bond interactions in galectin-3
to be published
|
|
6RSJ
 
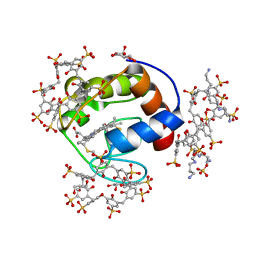 | |
6RSP
 
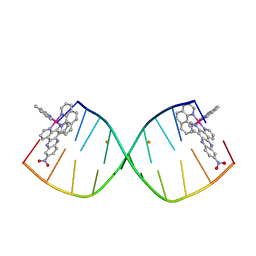 | |
5NBN
 
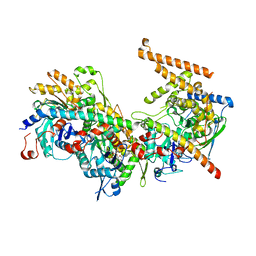 | | Crystal structure of the Arp4-N-actin-Arp8-Ino80HSA module of INO80 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-like protein ARP8, ... | | Authors: | Knoll, K.R, Eustermann, S, Hopfner, K.P. | | Deposit date: | 2017-03-02 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The nuclear actin-containing Arp8 module is a linker DNA sensor driving INO80 chromatin remodeling.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6RT8
 
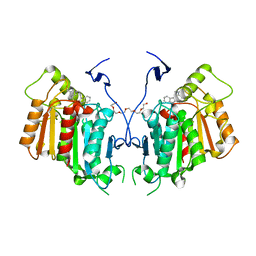 | | Structure of catharanthine synthase - an alpha-beta hydrolase from Catharanthus roseus with a cleaviminium intermediate bound | | Descriptor: | 18-carboxymethoxy-cleaviminium, Catharanthine synthase, HEXAETHYLENE GLYCOL | | Authors: | Caputi, L, Franke, J, Bussey, K, Farrow, S.C, Curcino Vieira, I.J, Stevenson, C.E.M, Lawson, D.M, O'Connor, S.E. | | Deposit date: | 2019-05-22 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural basis of cycloaddition in biosynthesis of iboga and aspidosperma alkaloids.
Nat.Chem.Biol., 16, 2020
|
|
5NCI
 
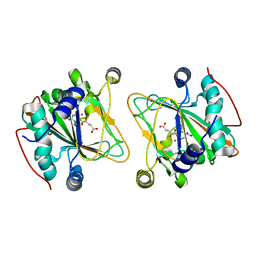 | | GriE in complex with cobalt, alpha-ketoglutarate and l-leucine | | Descriptor: | 2-OXOGLUTARIC ACID, COBALT (II) ION, LEUCINE, ... | | Authors: | Lukat, P, Blankenfeldt, W, Mueller, R. | | Deposit date: | 2017-03-05 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Biosynthesis of methyl-proline containing griselimycins, natural products with anti-tuberculosis activity.
Chem Sci, 8, 2017
|
|
6S1L
 
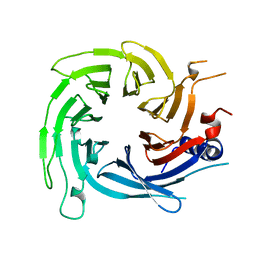 | |
6S1Z
 
 | | Crystal structure of Anopheles gambiae AnoACE2 in complex with fosinoprilat | | Descriptor: | Angiotensin-converting enzyme, ZINC ION, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R, Cashman, J.S. | | Deposit date: | 2019-06-19 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of angiotensin-converting enzyme from Anopheles gambiae in its native form and with a bound inhibitor.
Biochem.J., 476, 2019
|
|
5NMR
 
 | | Monomeric mouse Sortilin extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Leloup, N.O.L, Loessl, P, Meijer, D.H.M, Heck, A.J.R, Thies-Weesie, D.M.E, Janssen, B.J.C. | | Deposit date: | 2017-04-07 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Low pH-induced conformational change and dimerization of sortilin triggers endocytosed ligand release.
Nat Commun, 8, 2017
|
|
5NN4
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with N-acetyl-cysteine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roig-Zamboni, V, Cobucci-Ponzano, B, Iacono, R, Ferrara, M.C, Germany, S, Parenti, G, Bourne, Y, Moracci, M. | | Deposit date: | 2017-04-08 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of human lysosomal acid alpha-glucosidase-a guide for the treatment of Pompe disease.
Nat Commun, 8, 2017
|
|
