6UML
 
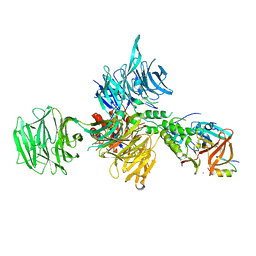 | | Structural Basis for Thalidomide Teratogenicity Revealed by the Cereblon-DDB1-SALL4-Pomalidomide Complex | | Descriptor: | DNA damage-binding protein 1, Protein cereblon, S-Pomalidomide, ... | | Authors: | Clayton, T.L, Matyskiela, M.E, Pagarigan, B.E, Tran, E.T, Chamberlain, P.P. | | Deposit date: | 2019-10-09 | | Release date: | 2020-04-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Crystal structure of the SALL4-pomalidomide-cereblon-DDB1 complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4NKG
 
 | | Crystal structure of SspH1 LRR domain in complex PKN1 HR1b domain | | Descriptor: | E3 ubiquitin-protein ligase sspH1, HEXANE-1,6-DIOL, Serine/threonine-protein kinase N1 | | Authors: | Keszei, A.F.A, Xiaojing, T, Mccormick, C, Zeqiraj, E, Rohde, J.R, Tyers, M, Sicheri, F. | | Deposit date: | 2013-11-12 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of an SspH1-PKN1 Complex Reveals the Basis for Host Substrate Recognition and Mechanism of Activation for a Bacterial E3 Ubiquitin Ligase.
Mol.Cell.Biol., 34, 2014
|
|
1JZL
 
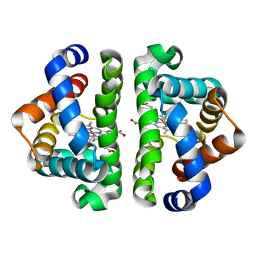 | | Crystal structure of Sapharca inaequivalvis HbI, I114M mutant ligated to carbon monoxide. | | Descriptor: | CARBON MONOXIDE, GLOBIN I - ARK SHELL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Knapp, J.E, Gibson, Q.H, Cushing, L, Royer Jr, W.E. | | Deposit date: | 2001-09-16 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Restricting the Ligand-Linked Heme Movement in Scapharca Dimeric Hemoglobin Reveals Tight Coupling between Distal and Proximal
Contributions to Cooperativity.
Biochemistry, 40, 2001
|
|
1MM6
 
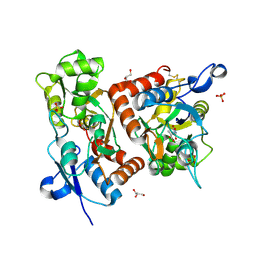 | | crystal structure of the GluR2 ligand binding core (S1S2J) in complex with quisqualate in a non zinc crystal form at 2.15 angstroms resolution | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, GLUTAMATE RECEPTOR 2, GLYCEROL, ... | | Authors: | Jin, R, Horning, M, Mayer, M.L, Gouaux, E. | | Deposit date: | 2002-09-03 | | Release date: | 2003-02-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mechanism of activation and selectivity in a ligand-gated ion channel: Structural and functional studies of GluR2 and quisqualate
Biochemistry, 41, 2002
|
|
2HCD
 
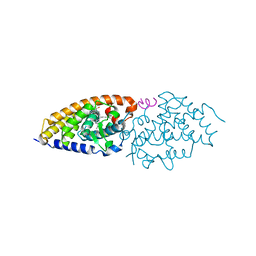 | | Crystal structure of the ligand binding domain of the Vitamin D nuclear receptor in complex with Gemini and a coactivator peptide | | Descriptor: | 21-NOR-9,10-SECOCHOLESTA-5,7,10(19)-TRIENE-1,3,25-TRIOL, 20-(4-HYDROXY-4-METHYLPENTYL)-, (1A,3B,5Z,7E), ... | | Authors: | Ciesielski, F, Rochel, N, Moras, D. | | Deposit date: | 2006-06-16 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Adaptability of the Vitamin D nuclear receptor to the synthetic ligand Gemini: remodelling the LBP with one side chain rotation
J.Steroid Biochem.Mol.Biol., 103, 2007
|
|
2D32
 
 | | Crystal Structure of Michaelis Complex of gamma-Glutamylcysteine Synthetase | | Descriptor: | CYSTEINE, GLUTAMIC ACID, Glutamate--cysteine ligase, ... | | Authors: | Hibi, T, Nakayama, M, Nii, H, Kurokawa, Y, Katano, H, Oda, J. | | Deposit date: | 2005-09-25 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of efficient coupling peptide ligation and ATP hydrolysis by gamma-gluatamylcysteine synthetase
To be Published
|
|
2PFK
 
 | |
2NQZ
 
 | | Trna-guanine transglycosylase (TGT) mutant in complex with 7-deaza-7-aminomethyl-guanine | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Tidten, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2006-11-01 | | Release date: | 2007-11-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor
Plos One, 8, 2013
|
|
2NSO
 
 | | Trna-gunanine-transglycosylase (TGT) mutant Y106F, C158V, A232S, V233G- APO-Structure | | Descriptor: | GLYCEROL, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Tidten, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2006-11-06 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor.
Plos One, 8, 2013
|
|
2HVC
 
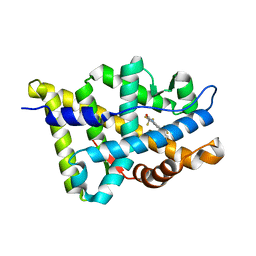 | | The Crystal Structure of Ligand-binding Domain (LBD) of human Androgen Receptor in Complex with a selective modulator LGD2226 | | Descriptor: | 6-[BIS(2,2,2-TRIFLUOROETHYL)AMINO]-4-(TRIFLUOROMETHYL)QUINOLIN-2(1H)-ONE, Androgen receptor | | Authors: | Wang, F, Liu, X.-Q, Li, H, Liang, K.-N, Miner, J.N, Hong, M, Kallel, E.A, van Oeveren, A, Zhi, L, Jiang, T. | | Deposit date: | 2006-07-28 | | Release date: | 2007-07-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the ligand-binding domain (LBD) of human androgen receptor in complex with a selective modulator LGD2226
ACTA CRYSTALLOGR.,SECT.F, 62, 2006
|
|
1ZNE
 
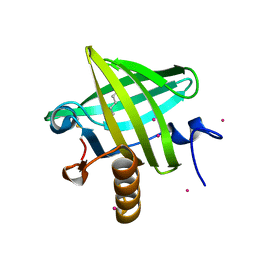 | | Strong Solute-Solute Dispersive Interactions in a Protein-Ligand Complex | | Descriptor: | CADMIUM ION, HEXAN-1-OL, Major Urinary Protein | | Authors: | Malham, R, Johnstone, S, Bingham, R.J, Barratt, E, Phillips, S.E, Laughton, C.A, Homans, S.W. | | Deposit date: | 2005-05-11 | | Release date: | 2005-12-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Strong Solute-Solute Dispersive Interactions in a Protein-Ligand Complex.
J.Am.Chem.Soc., 127, 2005
|
|
6TAY
 
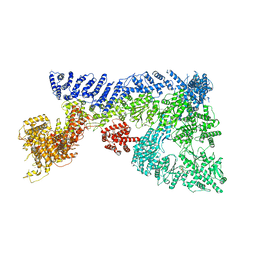 | | Mouse RNF213 mutant R4753K modeling the Moyamoya-disease-related Human variant R4810K | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNF213,E3 ubiquitin-protein ligase RNF213,E3 ubiquitin-protein ligase RNF213, ... | | Authors: | Ahel, J, Meinhart, A, Haselbach, D, Clausen, T. | | Deposit date: | 2019-10-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Moyamoya disease factor RNF213 is a giant E3 ligase with a dynein-like core and a distinct ubiquitin-transfer mechanism.
Elife, 9, 2020
|
|
8S6F
 
 | | A Structural Investigation of the Interaction between a GC-376-Based Peptidomimetic PROTAC and Its Precursor with the Viral Main Protease of Coxsackievirus B3 | | Descriptor: | Protease 3C, methyl (4~{S})-4-[[(2~{S})-4-methyl-2-(phenylmethoxycarbonylamino)pentanoyl]amino]-5-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]pentanoate | | Authors: | De Santis, A, Grifagni, D, Orsetti, A, Lenci, E, Rosato, A, D'Onofrio, M, Trabocchi, A, Ciofi-Baffoni, S, Cantini, F, Calderone, V. | | Deposit date: | 2024-02-27 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A Structural Investigation of the Interaction between a GC-376-Based Peptidomimetic PROTAC and Its Precursor with the Viral Main Protease of Coxsackievirus B3.
Biomolecules, 14, 2024
|
|
5MVB
 
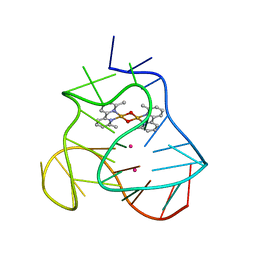 | | Solution structure of a human G-Quadruplex hybrid-2 form in complex with a Gold-ligand. | | Descriptor: | DNA (26-MER), POTASSIUM ION, bis(m2-Oxo)-bis(2-methyl-2,2'-bipyridine)-di-gold(iii) | | Authors: | Wirmer-Bartoschek, J, Jonker, H.R.A, Bendel, L.E, Gruen, T, Bazzicalupi, C, Messori, L, Gratteri, P, Schwalbe, H. | | Deposit date: | 2017-01-16 | | Release date: | 2017-05-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of a Ligand/Hybrid-2-G-Quadruplex Complex Reveals Rearrangements that Affect Ligand Binding.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
3KNP
 
 | |
2ZDH
 
 | |
3KO7
 
 | |
2ZDQ
 
 | |
3KOD
 
 | |
6TAX
 
 | | Mouse RNF213 wild type protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNF213,E3 ubiquitin-protein ligase RNF213,E3 ubiquitin-protein ligase RNF213, ... | | Authors: | Ahel, J, Meinhart, A, Haselbach, D, Clausen, T. | | Deposit date: | 2019-10-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Moyamoya disease factor RNF213 is a giant E3 ligase with a dynein-like core and a distinct ubiquitin-transfer mechanism.
Elife, 9, 2020
|
|
6R3K
 
 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with low molecular mass inhibitor | | Descriptor: | (2~{S},4~{R})-1-[[5-chloranyl-2-[(3-cyanophenyl)methoxy]-4-[[3-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-methyl-phenyl]methoxy]phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, Programmed cell death 1 ligand 1 | | Authors: | Zak, K.M, Grudnik, P, Skalniak, L, Dubin, G, Holak, T.A. | | Deposit date: | 2019-03-20 | | Release date: | 2019-04-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Terphenyl-Based Small-Molecule Inhibitors of Programmed Cell Death-1/Programmed Death-Ligand 1 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7P2K
 
 | | Solution NMR Structure of Arginine to Cysteine mutant of Arkadia RING domain. | | Descriptor: | E3 ubiquitin-protein ligase Arkadia, ZINC ION | | Authors: | Raptis, V, Marousis, K.D, Birkou, M, Bentrop, D, Episkopou, V, Spyroulias, G.A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Impact of a Single Nucleotide Polymorphism on the 3D Protein Structure and Ubiquitination Activity of E3 Ubiquitin Ligase Arkadia.
Front Mol Biosci, 9, 2022
|
|
4IC2
 
 | | Crystal structure of the XIAP RING domain | | Descriptor: | E3 ubiquitin-protein ligase XIAP, NICKEL (II) ION, ZINC ION | | Authors: | Linke, K, Nakatani, Y, Day, C.L. | | Deposit date: | 2012-12-09 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Regulation of ubiquitin transfer by XIAP, a dimeric RING E3 ligase
Biochem.J., 450, 2013
|
|
1IIW
 
 | |
8YAQ
 
 | |
