5DCO
 
 | |
7LMD
 
 | | SARS-CoV-2 3CLPro in complex with 2-(benzotriazol-1-yl)-N-[4-(1H-pyrazol-4-yl)phenyl]-N-(3-thienylmethyl)acetamide | | Descriptor: | 2-(benzotriazol-1-yl)-~{N}-[4-(1~{H}-pyrazol-4-yl)phenyl]-~{N}-(thiophen-3-ylmethyl)ethanamide, 3C-like proteinase | | Authors: | Goins, C.M, Arya, T, Macdonald, J.D, Stauffer, S.R. | | Deposit date: | 2021-02-05 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-Based Optimization of ML300-Derived, Noncovalent Inhibitors Targeting the Severe Acute Respiratory Syndrome Coronavirus 3CL Protease (SARS-CoV-2 3CL pro ).
J.Med.Chem., 65, 2022
|
|
8WDF
 
 | | Chemoreceptor PilJ from Pseudomonas aeruginosa PA14 | | Descriptor: | Chemotaxis chemoreceptor PilJ | | Authors: | Cui, R, Li, D.F. | | Deposit date: | 2023-09-15 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.996 Å) | | Cite: | The ligand binding domain of a type IV pilus chemoreceptor PilJ has a different fold from that of another PilJ-type receptor McpN.
Biochem.Biophys.Res.Commun., 706, 2024
|
|
3DQD
 
 | |
4HEL
 
 | |
1OBW
 
 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE | | Descriptor: | INORGANIC PYROPHOSPHATASE, MAGNESIUM ION | | Authors: | Oganessyan, V.Yu, Harutyunyan, E.H, Avaeva, S.M, Oganessyan, N.N, Mather, T, Huber, R. | | Deposit date: | 1996-10-09 | | Release date: | 1997-09-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of holo inorganic pyrophosphatase from Escherichia coli at 1.9 A resolution. Mechanism of hydrolysis.
Biochemistry, 36, 1997
|
|
7R1V
 
 | |
4L5Z
 
 | |
4H7M
 
 | |
5EF3
 
 | | RADIATION DAMAGE TO THE TRAP-RNA COMPLEX: DOSE (DWD) 25.0 MGy | | Descriptor: | (GAGUU)10GAG 53-NUCLEOTIDE RNA, TRYPTOPHAN, Transcription attenuation protein MtrB | | Authors: | Bury, C.S, McGeehan, J.E, Garman, E.F, Shevtsov, M.B. | | Deposit date: | 2015-10-23 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | RNA protects a nucleoprotein complex against radiation damage.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4H2G
 
 | | Human ecto-5'-nucleotidase (CD73): crystal form II (open) in complex with adenosine | | Descriptor: | 5'-nucleotidase, ADENOSINE, CALCIUM ION, ... | | Authors: | Straeter, N, Knapp, K.M, Zebisch, M, Pippel, J. | | Deposit date: | 2012-09-12 | | Release date: | 2012-11-28 | | Last modified: | 2012-12-26 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of the Human Ecto-5'-Nucleotidase (CD73): Insights into the Regulation of Purinergic Signaling.
Structure, 20, 2012
|
|
7LME
 
 | | SARS-CoV-2 3CLPro in complex with N-[4-[[2-(benzotriazol-1-yl)acetyl]-(3-thienylmethyl)amino]phenyl]cyclopropanecarboxamide | | Descriptor: | 3C-like proteinase, ~{N}-[4-[2-(benzotriazol-1-yl)ethanoyl-(thiophen-3-ylmethyl)amino]phenyl]cyclopropanecarboxamide | | Authors: | Goins, C.M, Arya, T, Macdonald, J.D, Stauffer, S.R. | | Deposit date: | 2021-02-05 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Optimization of ML300-Derived, Noncovalent Inhibitors Targeting the Severe Acute Respiratory Syndrome Coronavirus 3CL Protease (SARS-CoV-2 3CL pro ).
J.Med.Chem., 65, 2022
|
|
4L65
 
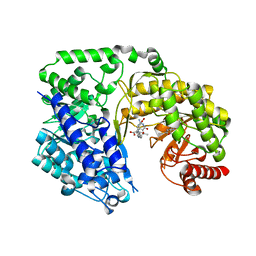 | | Crystal structure of the Candida albicans Methionine Synthase in complex with 5-Methyl-Tetrahydrofolate and Methionine | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, METHIONINE, ... | | Authors: | Ubhi, D, Robertus, J.D. | | Deposit date: | 2013-06-12 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural analysis of a fungal methionine synthase with substrates and inhibitors.
J.Mol.Biol., 426, 2014
|
|
1OAQ
 
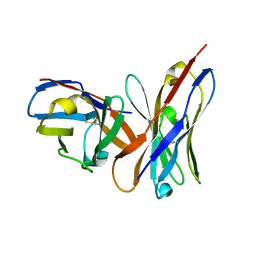 | |
3FZJ
 
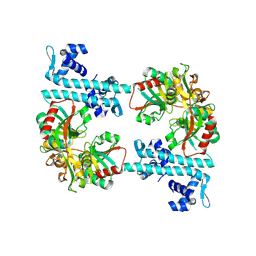 | | TsaR low resolution crystal structure, tetragonal form | | Descriptor: | LysR type regulator of tsaMBCD | | Authors: | Monferrer, D, Tralau, T, Kertesz, M.A, Dix, I, Kikhney, A.G, Svergun, D.I, Uson, I. | | Deposit date: | 2009-01-26 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (7.1 Å) | | Cite: | Structural studies on the full-length LysR-type regulator TsaR from Comamonas testosteroni T-2 reveal a novel open conformation of the tetrameric LTTR fold
Mol.Microbiol., 75, 2010
|
|
2WZS
 
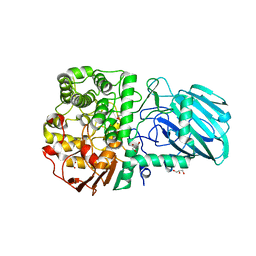 | | Structure of the Family GH92 Inverting Mannosidase BT3990 from Bacteroides thetaiotaomicron VPI-5482 in complex with Mannoimidazole | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Zhu, Y, Suits, M.D.L, Thompson, A, Chavan, S, Dinev, Z, Dumon, C, Smith, N, Moremen, K.W, Xiang, Y, Siriwardena, A, Williams, S.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2009-12-02 | | Release date: | 2009-12-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanistic Insights Into a Ca2+-Dependent Family of A-Mannosidases in a Human Gut Symbiont.
Nat.Chem.Biol., 6, 2010
|
|
4LLG
 
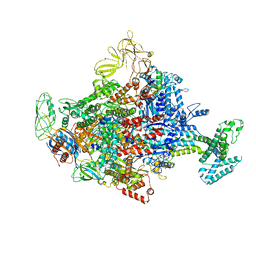 | | Crystal Structure Analysis of the E.coli holoenzyme/Gp2 complex | | Descriptor: | Bacterial RNA polymerase inhibitor, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Bae, B, Darst, S.A. | | Deposit date: | 2013-07-09 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.7936 Å) | | Cite: | Phage T7 Gp2 inhibition of Escherichia coli RNA polymerase involves misappropriation of sigma 70 domain 1.1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7R3W
 
 | |
2ZMN
 
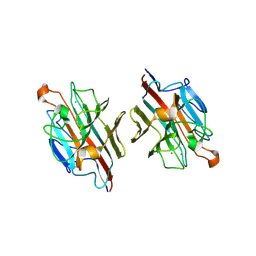 | | Crystal Structure of basic winged bean lectin in complex with Gal-alpha- 1,6 Glc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Basic agglutinin, CALCIUM ION, ... | | Authors: | Kulkarni, K.A, Katiyar, S, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2008-04-19 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and sugar-specificity of basic winged-bean lectin: structures of new disaccharide complexes and a comparative study with other known disaccharide complexes of the lectin.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2WNH
 
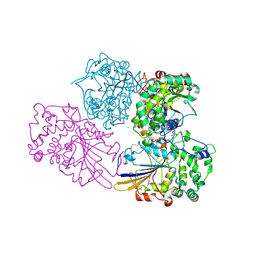 | | Crystal Structure Analysis of Klebsiella sp ASR1 Phytase | | Descriptor: | 3-PHYTASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Bohm, K, Mueller, J.J, Heinemann, U. | | Deposit date: | 2009-07-09 | | Release date: | 2010-04-28 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of Klebsiella Sp. Asr1 Phytase Suggests Substrate Binding to a Preformed Active Site that Meets the Requirements of a Plant Rhizosphere Enzyme.
FEBS J., 277, 2010
|
|
7L8G
 
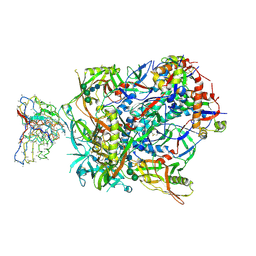 | | BG505 SOSIP.v5.2(7S) in complex with the polyclonal Fab pAbC-3 from animal Rh.33172 (Wk38 time point) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Antanasijevic, A, Sewall, L.M, Rantalainen, K, Ward, A.B. | | Deposit date: | 2020-12-31 | | Release date: | 2021-08-04 | | Last modified: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Polyclonal antibody responses to HIV Env immunogens resolved using cryoEM.
Nat Commun, 12, 2021
|
|
5EEX
 
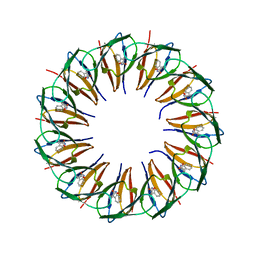 | | RADIATION DAMAGE TO THE TRAP-RNA COMPLEX: DOSE (DWD) 9.02 MGy | | Descriptor: | (GAGUU)10GAG 53-NUCLEOTIDE RNA, TRYPTOPHAN, Transcription attenuation protein MtrB | | Authors: | Bury, C.S, McGeehan, J.E, Garman, E.F, Shevtsov, M.B. | | Deposit date: | 2015-10-23 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | RNA protects a nucleoprotein complex against radiation damage.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
2ZO5
 
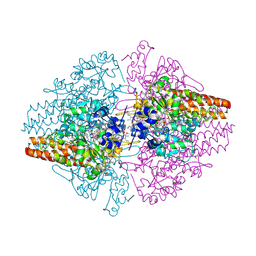 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in a complex with azide | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AZIDE ION, ... | | Authors: | Polyakov, K.M, Boyko, K.M, Slutsky, A, Tikhonova, T.V, Antipov, A.N, Zvyagilskaya, R.A, Popov, A.N, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2008-05-05 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution structural analysis of a novel octaheme cytochrome c nitrite reductase from the haloalkaliphilic bacterium Thioalkalivibrio nitratireducens
J.Mol.Biol., 389, 2009
|
|
2Z9K
 
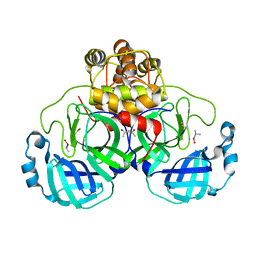 | | Complex structure of SARS-CoV 3C-like protease with JMF1600 | | Descriptor: | (dimethylamino)(hydroxy)zinc', 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Lee, C.C, Wang, A.H. | | Deposit date: | 2007-09-20 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of mercury- and zinc-conjugated complexes as SARS-CoV 3C-like protease inhibitors.
Febs Lett., 581, 2007
|
|
4LAA
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L36H at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Wheeler, E.L, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B, Robinson, A.C. | | Deposit date: | 2013-06-19 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS L36H at cryogenic temperature
To be Published
|
|
