3KM6
 
 | |
4V4S
 
 | | Crystal structure of the whole ribosomal complex. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petry, S, Brodersen, D.E, Murphy IV, F.V, Dunham, C.M, Selmer, M, Tarry, M.J, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2005-10-12 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (6.76 Å) | | Cite: | Crystal Structures of the Ribosome in Complex with Release Factors RF1 and RF2 Bound to a Cognate Stop Codon.
Cell(Cambridge,Mass.), 123, 2005
|
|
4V5F
 
 | | The structure of the ribosome with elongation factor G trapped in the post-translocational state | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, Y.-G, Selmer, M, Dunham, C.M, Weixlbaumer, A, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-09-01 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The structure of the ribosome with elongation factor G trapped in the posttranslocational state.
Science, 326, 2009
|
|
1QX5
 
 | |
4V5N
 
 | | tRNA translocation on the 70S ribosome: the post- translocational translocation intermediate TI(POST) | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ratje, A.H, Loerke, J, Mikolajka, A, Bruenner, M, Hildebrand, P.W, Starosta, A.L, Doenhoefer, A, Connell, S.R, Fucini, P, Mielke, T, Whitford, P.C, Onuchic, J.N, Yu, Y, Sanbonmatsu, K.Y, Hartmann, R.K, Penczek, P.A, Wilson, D.N, Spahn, C.M.T. | | Deposit date: | 2010-10-21 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Head Swivel on the Ribosome Facilitates Translocation by Means of Intra-Subunit tRNA Hybrid Sites.
Nature, 468, 2010
|
|
3KPY
 
 | | Crystal Structure of hPNMT in Complex AdoHcy and 6-Chlorooxindole | | Descriptor: | 6-chloro-1,3-dihydro-2H-indol-2-one, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
1R0N
 
 | | Crystal Structure of Heterodimeric Ecdsyone receptor DNA binding complex | | Descriptor: | Ecdsyone Response Element, Ecdysone Response Element, Ecdysone receptor, ... | | Authors: | Devarakonda, S, Harp, J.M, Kim, Y, Ozyhar, A, Rastinejad, F. | | Deposit date: | 2003-09-22 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the heterodimeric Ecdysone Receptor DNA-binding complex
Embo J., 22, 2003
|
|
3KK2
 
 | | HIV-1 reverse transcriptase-DNA complex with dATP bound in the nucleotide binding site | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*A*TP*GP*GP*TP*GP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3', 5'-D(*AP*CP*A*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*(DOC))-3', ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2009-11-04 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Visualizing the molecular interactions of a nucleotide analog, GS-9148, with HIV-1 reverse transcriptase-DNA complex.
J.Mol.Biol., 397, 2010
|
|
1QKI
 
 | | X-RAY STRUCTURE OF HUMAN GLUCOSE 6-PHOSPHATE DEHYDROGENASE (VARIANT CANTON R459L) COMPLEXED WITH STRUCTURAL NADP+ | | Descriptor: | GLUCOSE-6-PHOSPHATE 1-DEHYDROGENASE, GLYCEROL, GLYCOLIC ACID, ... | | Authors: | Au, S.W.N, Gover, S, Lam, V.M.S, Adams, M.J. | | Deposit date: | 1999-07-20 | | Release date: | 2000-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human Glucose-6-Phosphate Dehydrogenase: The Crystal Structure Reveals a Structural Nadp+ Molecule and Provides Insights Into Enzyme Deficiency
Structure, 8, 2000
|
|
3KQV
 
 | | Crystal Structure of hPNMT in Complex AdoHcy and Formanilide | | Descriptor: | FORMANILIDE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
1QMB
 
 | | Cleaved alpha-1-antitrypsin polymer | | Descriptor: | ALPHA-1-ANTITRYPSIN | | Authors: | Huntington, J.A, Pannu, N.S, Hazes, B, Read, R.J, Lomas, D.A, Carrell, R.W. | | Deposit date: | 1999-09-24 | | Release date: | 2000-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A 2.6A Structure of a Serpin Polymer and Implications for Conformational Disease
J.Mol.Biol., 293, 1999
|
|
1QLX
 
 | | Human prion protein | | Descriptor: | PRION PROTEIN | | Authors: | Zahn, R, Liu, A, Luhrs, T, Wuthrich, K. | | Deposit date: | 1999-09-17 | | Release date: | 1999-12-16 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the Human Prion Protein
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1R55
 
 | | Crystal structure of the catalytic domain of human ADAM 33 | | Descriptor: | (2S,3R)-N~4~-[(1S)-2,2-dimethyl-1-(methylcarbamoyl)propyl]-N~1~,2-dihydroxy-3-(2-methylpropyl)butanediamide, ADAM 33, CALCIUM ION, ... | | Authors: | Orth, P, Reichert, P, Wang, W, Prosise, W.W, Yarosh-Tomaine, T, Hammond, G, Xiao, L, Mirza, U.A, Zou, J, Strickland, C, Taremi, S.S, Le, H.V, Madison, V. | | Deposit date: | 2003-10-09 | | Release date: | 2004-10-12 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structre of the catalytic domain of human ADAM33
J.Mol.Biol., 335, 2004
|
|
5GHT
 
 | | DNA replication protein | | Descriptor: | SsDNA-specific exonuclease | | Authors: | Oyama, T. | | Deposit date: | 2016-06-20 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.795 Å) | | Cite: | Atomic structure of an archaeal GAN suggests its dual roles as an exonuclease in DNA repair and a CMG component in DNA replication.
Nucleic Acids Res., 44, 2016
|
|
8U7F
 
 | | Crystal structure of CIB_12 beta-galactosidase from Cuniculiplasma divulgatum | | Descriptor: | CIB_12 Beta-galactosidase, GLYCEROL | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Yakunin, A, Golyshin, P, Savchenko, A. | | Deposit date: | 2023-09-15 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of CIB_12 beta-galactosidase from Cuniculiplasma divulgatum
To Be Published
|
|
8U7G
 
 | |
3I5Q
 
 | | Nup170(aa1253-1502) at 2.2 A, S.cerevisiae | | Descriptor: | Nucleoporin NUP170 | | Authors: | Whittle, J.R.R, Schwartz, T.U. | | Deposit date: | 2009-07-06 | | Release date: | 2009-08-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Architectural nucleoporins Nup157/170 and Nup133 are structurally related and descend from a second ancestral element.
J.Biol.Chem., 284, 2009
|
|
7NKX
 
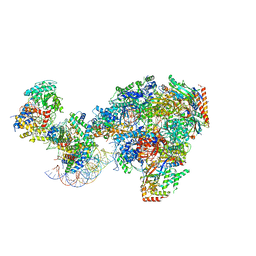 | | RNA polymerase II-Spt4/5-nucleosome-Chd1 structure | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromatin elongation factor SPT4, ... | | Authors: | Farnung, L, Ochmann, M, Engeholm, M, Cramer, P. | | Deposit date: | 2021-02-19 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of nucleosome transcription mediated by Chd1 and FACT.
Nat.Struct.Mol.Biol., 28, 2021
|
|
8V4T
 
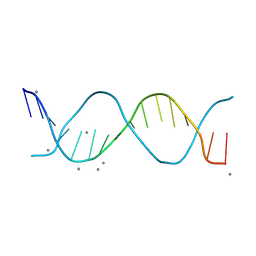 | |
8V58
 
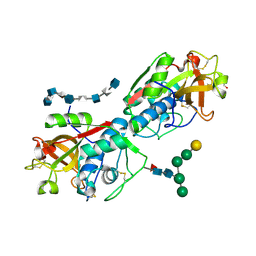 | | Complex of murine cathepsin K with bound heparan sulfate 12mer | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, ... | | Authors: | Pedersen, L.C, Xu, D, Krahn, J.M. | | Deposit date: | 2023-11-30 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Heparan sulfate selectively inhibits the collagenase activity of cathepsin K.
Matrix Biol., 129, 2024
|
|
8V57
 
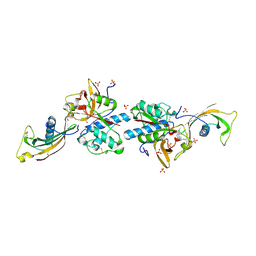 | | Complex of murine cathepsin K with bound cystatin C inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cathepsin K, ... | | Authors: | Pedersen, L.C, Xu, D. | | Deposit date: | 2023-11-30 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Heparan sulfate selectively inhibits the collagenase activity of cathepsin K.
Matrix Biol., 129, 2024
|
|
8VVA
 
 | |
8VS5
 
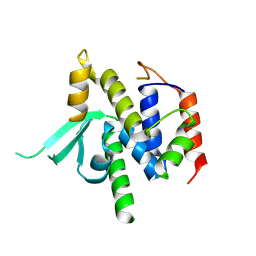 | | Structure of catalytic domain of telomere resolvase, ResT, from Borrelia garinii | | Descriptor: | Telomere resolvase ResT | | Authors: | Semper, C, Savchenko, A, Watanabe, N, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-01-23 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Structure analysis of the telomere resolvase from the Lyme disease spirochete Borrelia garinii reveals functional divergence of its C-terminal domain.
Nucleic Acids Res., 52, 2024
|
|
8VJ1
 
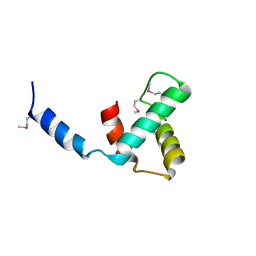 | | Structure of C-terminal domain of telomere resolvase, ResT, from Borrelia garinii | | Descriptor: | Telomere resolvase ResT | | Authors: | Semper, C, Savchenko, A, Watanabe, N, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure analysis of the telomere resolvase from the Lyme disease spirochete Borrelia garinii reveals functional divergence of its C-terminal domain.
Nucleic Acids Res., 52, 2024
|
|
3I4R
 
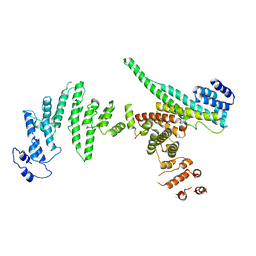 | | Nup107(aa658-925)/Nup133(aa517-1156) complex, H.sapiens | | Descriptor: | Nuclear pore complex protein Nup107, Nuclear pore complex protein Nup133 | | Authors: | Whittle, J.R.R, Schwartz, T.U. | | Deposit date: | 2009-07-02 | | Release date: | 2009-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | Architectural nucleoporins Nup157/170 and Nup133 are structurally related and descend from a second ancestral element.
J.Biol.Chem., 284, 2009
|
|
