4MLY
 
 | | Disulfide isomerase from multidrug resistance IncA/C related integrative and conjugative elements in oxidized state (P21 space group) | | Descriptor: | 1,3-BUTANEDIOL, DsbP | | Authors: | Premkumar, L, Kurth, F, Neyer, S, Martin, J.L. | | Deposit date: | 2013-09-06 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.207 Å) | | Cite: | The Multidrug Resistance IncA/C Transferable Plasmid Encodes a Novel Domain-swapped Dimeric Protein-disulfide Isomerase.
J.Biol.Chem., 289, 2014
|
|
5K6J
 
 | | Human Phospodiesterase 4B in complex with pyridyloxy-benzoxaborole based inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 6-[[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1(6),2,4-trien-3-yl]oxy]-5-chloranyl-2-(4-oxidanylidenepentoxy)pyridine-3-carbonitrile, MAGNESIUM ION, ... | | Authors: | Rock, F.L, Zhou, Y, Sullivan, D. | | Deposit date: | 2016-05-24 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | To be published
To Be Published
|
|
5ECQ
 
 | | Crystal Structure of FIN219-FIP1 complex with JA, VAL and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLUTATHIONE, Glutathione S-transferase U20, ... | | Authors: | Chen, C.Y, Cheng, Y.S. | | Deposit date: | 2015-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural basis of jasmonate-amido synthetase FIN219 in complex with glutathione S-transferase FIP1 during the JA signal regulation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2GUE
 
 | |
4PXO
 
 | | Crystal structure of Maleylacetoacetate isomerase from Methylobacteriu extorquens AM1 WITH BOUND MALONATE AND GSH (TARGET EFI-507068) | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, MALONIC ACID, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-03-24 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of glutathione s-transferase zeta from Methylobacterium extorquens (TARGET EFI-507068)
To be Published
|
|
4HMQ
 
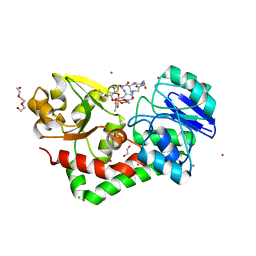 | | Crystal structure of streptococcus pneumoniae TIGR4 PiaA in complex with ferrichrome | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Cheng, W, Li, Q, Jiang, Y.-L, Chen, Y, Zhou, C.-Z. | | Deposit date: | 2012-10-18 | | Release date: | 2013-09-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Streptococcus pneumoniae PiaA and Its Complex with Ferrichrome Reveal Insights into the Substrate Binding and Release of High Affinity Iron Transporters
Plos One, 8, 2013
|
|
6JV9
 
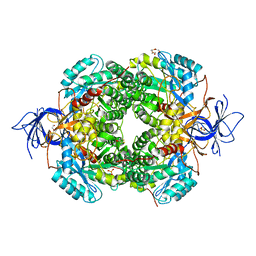 | | Crystal Structure of Human CRMP2 1-532, unmodified | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Dihydropyrimidinase-related protein 2, ... | | Authors: | Jiang, X, Ogawa, T, Hirokawa, N. | | Deposit date: | 2019-04-16 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Enhanced carbonyl stress induces irreversible multimerization of CRMP2 in schizophrenia pathogenesis.
Life Sci Alliance, 2, 2019
|
|
1WXS
 
 | | Solution Structure of Ufm1, a ubiquitin-fold modifier | | Descriptor: | Ubiquitin-fold Modifier 1 | | Authors: | Sasakawa, H, Sakata, E, Yamaguchi, Y, Komatsu, M, Tatsumi, K, Kominami, E, Tanaka, K, Kato, K. | | Deposit date: | 2005-02-01 | | Release date: | 2006-04-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of Ufm1, a ubiquitin-fold modifier 1
Biochem.Biophys.Res.Commun., 343, 2006
|
|
6K9O
 
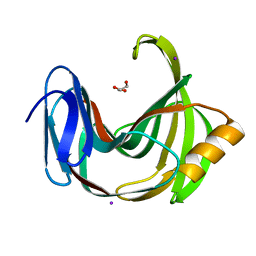 | | Crystal Structure Analysis of Protein | | Descriptor: | Endo-1,4-beta-xylanase 2, GLYCEROL, IODIDE ION | | Authors: | Li, C, Wan, Q. | | Deposit date: | 2019-06-17 | | Release date: | 2020-06-17 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Studying the Role of a Single Mutation of a Family 11 Glycoside Hydrolase Using High-Resolution X-ray Crystallography.
Protein J., 39, 2020
|
|
1BBH
 
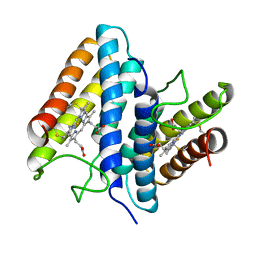 | |
5E7O
 
 | | Crystal structure of the perchlorate reductase PcrAB mutant W461E of PcrA from Azospira suillum PS | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DMSO reductase family type II enzyme, ... | | Authors: | Tsai, C.-L, Tainer, J.A. | | Deposit date: | 2015-10-12 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Perchlorate Reductase Is Distinguished by Active Site Aromatic Gate Residues.
J.Biol.Chem., 291, 2016
|
|
6KA6
 
 | | Crystal structure of plasmodium lysyl-tRNA synthetase in complex with a cladosporin derivative 1 | | Descriptor: | (3~{S})-3-[[(1~{S},3~{S})-3-methylcyclohexyl]methyl]-6,8-bis(oxidanyl)-3,4-dihydroisochromen-1-one, GLYCEROL, LYSINE, ... | | Authors: | Zhou, J, Fang, P. | | Deposit date: | 2019-06-21 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Atomic Resolution Analyses of Isocoumarin Derivatives for Inhibition of Lysyl-tRNA Synthetase.
Acs Chem.Biol., 15, 2020
|
|
4Q9N
 
 | | Crystal structure of Chlamydia trachomatis enoyl-ACP reductase (FabI) in complex with NADH and AFN-1252 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Enoyl-[acyl-carrier-protein] reductase [NADH], N-methyl-N-[(3-methyl-1-benzofuran-2-yl)methyl]-3-(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-3-yl)propanamide | | Authors: | Yao, J, Abdelrahman, Y, Robertson, R.M, Cox, J.V, Belland, R.J, White, S.W, Rock, C.O. | | Deposit date: | 2014-05-01 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Type II Fatty Acid Synthesis Is Essential for the Replication of Chlamydia trachomatis.
J.Biol.Chem., 289, 2014
|
|
7H72
 
 | | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with Z1562205518 (CHIKV_MacB-x0477) | | Descriptor: | 1-(2-hydroxyethyl)-1H-pyrazole-4-carboxamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Balcomb, B.H, Capkin, E, Chandran, A.V, Dolci, I, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Oliva, G, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain
To Be Published
|
|
3G56
 
 | | Structure of the macrolide biosensor protein, MphR(A) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Regulator of macrolide 2'-phosphotransferase I | | Authors: | Zheng, J, Sagar, V, Smolinsky, A, Bourke, C, LaRonde-LeBlanc, N, Cropp, T.A. | | Deposit date: | 2009-02-04 | | Release date: | 2009-03-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of the macrolide biosensor protein, MphR(A), with and without erythromycin
J.Mol.Biol., 387, 2009
|
|
2WL1
 
 | | Pyrin PrySpry domain | | Descriptor: | 1,2-ETHANEDIOL, PYRIN, THIOCYANATE ION | | Authors: | Weinert, C, Mittl, P.R, Gruetter, M.G. | | Deposit date: | 2009-06-19 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Crystal Structure of Human Pyrin B30.2 Domain: Implications for Mutations Associated with Familial Mediterranean Fever.
J.Mol.Biol., 394, 2009
|
|
3G7N
 
 | | Crystal Structure of a Triacylglycerol Lipase from Penicillium Expansum at 1.3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Lipase, PENTAETHYLENE GLYCOL, ... | | Authors: | Bian, C.B, Yuan, C, Chen, L.Q, Edward, J.M, Lin, L, Jiang, L.G, Huang, Z.X, Huang, M.D. | | Deposit date: | 2009-02-10 | | Release date: | 2010-02-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of a triacylglycerol lipase from Penicillium expansum at 1.3 A determined by sulfur SAD
Proteins, 78, 2010
|
|
4MSP
 
 | | Crystal structure of human peptidyl-prolyl cis-trans isomerase FKBP22 (aka FKBP14) containing two EF-hand motifs | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase FKBP14, ... | | Authors: | Boudko, S.P, Ishikawa, Y, Bachinger, H.P. | | Deposit date: | 2013-09-18 | | Release date: | 2013-12-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of human peptidyl-prolyl cis-trans isomerase FKBP22 containing two EF-hand motifs.
Protein Sci., 23, 2014
|
|
7E9R
 
 | |
7X2L
 
 | | Crystal structure of nanobody 3-2A2-4 with SARS-CoV-2 RBD | | Descriptor: | Nanobody 3-2A2-4, Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | Deposit date: | 2022-02-25 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
4MIK
 
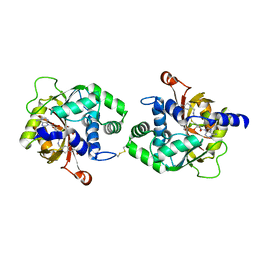 | | Crystal Structure of hPNMT in Complex with bisubstrate inhibitor (2R,3R,4S,5S)-2-(6-amino-9H-purin-9-yl)-5-(((2-(((7-nitro-1,2,3,4-tetrahydroisoquinolin-3-yl)methyl)amino)ethyl)thio)methyl)tetrahydrofuran-3,4-diol | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 9-{5-S-[2-({[(3S)-7-nitro-1,2,3,4-tetrahydroisoquinolin-3-yl]methyl}amino)ethyl]-5-thio-alpha-L-lyxofuranosyl}-9H-purin-6-amine, Phenylethanolamine N-methyltransferase | | Authors: | Bart, A.G, Scott, E.E. | | Deposit date: | 2013-08-31 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9506 Å) | | Cite: | To be published
To be Published
|
|
1RZT
 
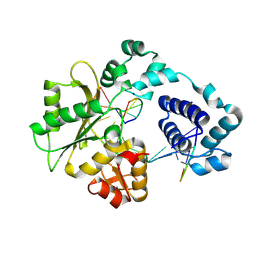 | | Crystal structure of DNA polymerase lambda complexed with a two nucleotide gap DNA molecule | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*CP*GP*GP*CP*AP*AP*CP*GP*CP*AP*C)-3', 5'-D(*GP*TP*GP*CP*G)-3', ... | | Authors: | Pedersen, L.C, Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Blanco, L, Kunkel, T.A. | | Deposit date: | 2003-12-29 | | Release date: | 2004-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural solution for the DNA polymerase lambda-dependent repair of DNA gaps with minimal homology.
Mol.Cell, 13, 2004
|
|
1PMY
 
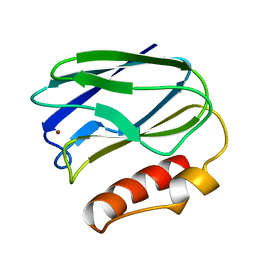 | | REFINED CRYSTAL STRUCTURE OF PSEUDOAZURIN FROM METHYLOBACTERIUM EXTORQUENS AM1 AT 1.5 ANGSTROMS RESOLUTION | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Inoue, T, Kai, Y, Harada, S, Kasai, N, Ohshiro, Y, Suzuki, S, Kohzuma, T, Tobari, J. | | Deposit date: | 1994-01-28 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Refined crystal structure of pseudoazurin from Methylobacterium extorquens AM1 at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1JV4
 
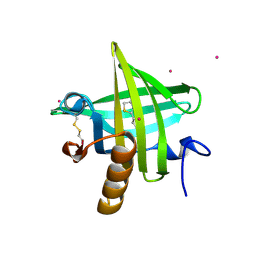 | | Crystal structure of recombinant major mouse urinary protein (rmup) at 1.75 A resolution | | Descriptor: | 2-(SEC-BUTYL)THIAZOLE, CADMIUM ION, Major urinary protein 2 | | Authors: | Kuser, P.R, Franzoni, L, Ferrari, E, Spisni, A, Polikarpov, I. | | Deposit date: | 2001-08-28 | | Release date: | 2001-12-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The X-ray structure of a recombinant major urinary protein at 1.75 A resolution. A comparative study of X-ray and NMR-derived structures.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
5TUO
 
 | | Crystal structure of the complex of Helicobacter pylori alpha-carbonic anhydrase with 5-amino-1,3,4-thiadiazole-2-sulfonamide inhibitor. | | Descriptor: | 5-AMINO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Alpha-carbonic anhydrase, CHLORIDE ION, ... | | Authors: | Modak, J.K, Roujeinikova, A. | | Deposit date: | 2016-11-06 | | Release date: | 2017-09-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Activity Relationship for Sulfonamide Inhibition of Helicobacter pylori alpha-Carbonic Anhydrase.
J. Med. Chem., 59, 2016
|
|
