8TVN
 
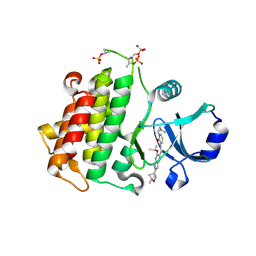 | | IRAK4 in complex with compound 23 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-{1-[(1R,2S)-2-fluorocyclopropyl]-2-oxo-1,2-dihydropyridin-3-yl}-2-[(1R,4r)-1-methyl-2-oxabicyclo[2.1.1]hexan-4-yl]-6-[(propan-2-yl)oxy]-2H-pyrazolo[3,4-b]pyridine-5-carboxamide | | Authors: | Metrick, C.M, Chodaparambil, J.V. | | Deposit date: | 2023-08-18 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Tiny Pocket Packs a Punch: Leveraging Pyridones for the Discovery of CNS-Penetrant Aza-indazole IRAK4 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
8TVM
 
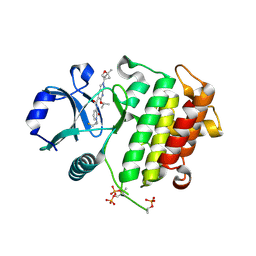 | | IRAK4 in complex with compound 24 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-{1-[(1R,2R)-2-fluorocyclopropyl]-2-oxo-1,2-dihydropyridin-3-yl}-2-[(1R,4r)-1-methyl-2-oxabicyclo[2.1.1]hexan-4-yl]-6-[(propan-2-yl)oxy]-2H-pyrazolo[3,4-b]pyridine-5-carboxamide | | Authors: | Metrick, C.M, Chodaparambil, J.V. | | Deposit date: | 2023-08-18 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Tiny Pocket Packs a Punch: Leveraging Pyridones for the Discovery of CNS-Penetrant Aza-indazole IRAK4 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
5MIK
 
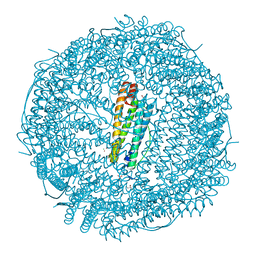 | | X-ray structure of carboplatin-encapsulated horse spleen apoferritin (rotating anode data) | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ferritin light chain, ... | | Authors: | Pontillo, N, Ferraro, G, Helliwell, J.R, Merlino, A. | | Deposit date: | 2016-11-28 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | X-ray Structure of the Carboplatin-Loaded Apo-Ferritin Nanocage.
ACS Med Chem Lett, 8, 2017
|
|
5MPK
 
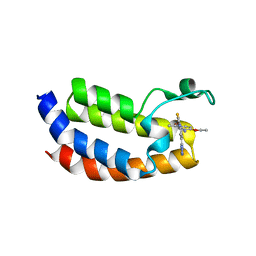 | | Crystal structure of CREBBP bromodomain complexed with DK19 | | Descriptor: | CREB-binding protein, ~{N}-(5-ethanoyl-2-ethoxy-phenyl)-3-(2~{H}-1,2,3,4-tetrazol-5-yl)-5-(1,3-thiazol-4-yl)benzamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2016-12-16 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
5MES
 
 | | MCL1 FAB COMPLEX IN COMPLEX WITH COMPOUND 29 | | Descriptor: | (5~{R},13~{S},17~{S})-5-[[4-chloranyl-3-(2-phenylethyl)phenyl]methyl]-13-[(4-chlorophenyl)methyl]-8-methyl-1,4,8,12,16-pentazatricyclo[15.8.1.0^{20,25}]hexacosa-20(25),21,23-triene-3,7,15,26-tetrone, Heavy Chain, Induced myeloid leukemia cell differentiation protein Mcl-1 homolog,Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | Authors: | Hargreaves, D. | | Deposit date: | 2016-11-16 | | Release date: | 2017-01-18 | | Last modified: | 2017-03-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
5MW4
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD7 [N-(3-(((R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl)(methyl)amino)propyl)-2-(3-(2-chloro-3-(2-methylpyridin-3-yl)benzo[b]thiophen-5-yl)ureido)acetamide] | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, N~2~-{[2-chloro-3-(2-methylpyridin-3-yl)-1-benzothiophen-5-yl]carbamoyl}-N-(3-{methyl[(3R)-1-(5H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]amino}propyl)glycinamide, ... | | Authors: | Be, C, Koch, E, Gaul, C, Stauffer, F, Moebitz, H, Scheufler, C. | | Deposit date: | 2017-01-18 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Discovery of Potent, Selective, and Structurally Novel Dot1L Inhibitors by a Fragment Linking Approach.
ACS Med Chem Lett, 8, 2017
|
|
8U7X
 
 | |
5MMG
 
 | | Crystal structure of CREBBP bromodomain complexed with UT07C | | Descriptor: | 1-[4-ethoxy-3-[(1-methylsulfonylindol-6-yl)amino]phenyl]ethanone, CREB-binding protein | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2016-12-09 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
8U7W
 
 | |
5MPN
 
 | | Crystal structure of CREBBP bromodomain complexed with FA26 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-ethoxy-3-[3-(2~{H}-1,2,3,4-tetrazol-5-yl)phenyl]phenyl]ethanone, CREB-binding protein | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2016-12-16 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
5MW3
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD1 [N6-(2,6-dichlorophenyl)-N6-(pent-2-yn-1-yl)quinoline-4,6-diamine] and inhibitor CPD2 [(R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-amine] | | Descriptor: | (3R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-amine, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Be, C, Koch, E, Gaul, C, Stauffer, F, Moebitz, H, Scheufler, C. | | Deposit date: | 2017-01-18 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of Potent, Selective, and Structurally Novel Dot1L Inhibitors by a Fragment Linking Approach.
ACS Med Chem Lett, 8, 2017
|
|
8VHL
 
 | | Structure of DHODH in Complex with Ligand 17 | | Descriptor: | ACETATE ION, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery of Alternative Binding Poses through Fragment-Based Identification of DHODH Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
8VHM
 
 | | Structure of DHODH in Complex with Fragment 2 | | Descriptor: | ACETATE ION, CHLORIDE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery of Alternative Binding Poses through Fragment-Based Identification of DHODH Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
5LUU
 
 | | Structure of the first bromodomain of BRD4 with a pyrazolo[4,3-c]pyridin fragment | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-phenyl-1,4,6,7-tetrahydropyrazolo[4,3-c]pyridin-5-yl)propan-1-one, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Knapp, S, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-11 | | Release date: | 2016-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Discovery of New Bromodomain Scaffolds by Biosensor Fragment Screening.
ACS Med Chem Lett, 7, 2016
|
|
1UJ6
 
 | | Crystal structure of Thermus thermophilus ribose-5-phosphate isomerase complexed with arabinose-5-phosphate | | Descriptor: | ARABINOSE-5-PHOSPHATE, CHLORIDE ION, ribose 5-phosphate isomerase | | Authors: | Hamada, K, Ago, H, Sugahara, M, Nodake, Y, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-26 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Oxyanion hole-stabilized stereospecific isomerization in ribose-5-phosphate isomerase (Rpi)
J.Biol.Chem., 278, 2003
|
|
5MVS
 
 | | Crystal structure of Dot1L in complex with adenosine and inhibitor CPD1 [N6-(2,6-dichlorophenyl)-N6-(pent-2-yn-1-yl)quinoline-4,6-diamine] | | Descriptor: | ADENOSINE, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Be, C, Koch, E, Gaul, C, Stauffer, F, Moebitz, H, Scheufler, C. | | Deposit date: | 2017-01-17 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of Potent, Selective, and Structurally Novel Dot1L Inhibitors by a Fragment Linking Approach.
ACS Med Chem Lett, 8, 2017
|
|
8W3W
 
 | | Crystal structure of IRAK4 in complex with compound 4 | | Descriptor: | 7-methoxy-1-{[(2S)-5-oxopyrrolidin-2-yl]methoxy}isoquinoline-6-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Knafels, J.D. | | Deposit date: | 2024-02-22 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.976 Å) | | Cite: | In Retrospect: Root-Cause Analysis of Structure-Activity Relationships in IRAK4 Inhibitor Zimlovisertib (PF-06650833).
Acs Med.Chem.Lett., 15, 2024
|
|
8W3X
 
 | | Crystal structure of IRAK4 in complex with compound 6 | | Descriptor: | 7-ethoxy-1-{[(2S)-5-oxopyrrolidin-2-yl]methoxy}isoquinoline-6-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Knafels, J.D. | | Deposit date: | 2024-02-22 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.765 Å) | | Cite: | In Retrospect: Root-Cause Analysis of Structure-Activity Relationships in IRAK4 Inhibitor Zimlovisertib (PF-06650833).
Acs Med.Chem.Lett., 15, 2024
|
|
8VXE
 
 | | Structure of p38 alpha (Mitogen-activated protein kinase 14) complexed with inhibitor 6 | | Descriptor: | (4M)-4-[3-(4-fluorophenyl)-1-methyl-1H-pyrazol-4-yl]-1H-pyrrolo[2,3-b]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Blaesse, M, Steinbacher, S, Shaffer, P.L, Sharma, S, Thompson, A.A. | | Deposit date: | 2024-02-04 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Optimization of Selective and Brain Penetrant CK1 delta Inhibitors for the Treatment of Circadian Disruptions.
Acs Med.Chem.Lett., 15, 2024
|
|
8VXD
 
 | |
8VXF
 
 | | Structure of Casein kinase I isoform delta (CK1d) complexed with inhibitor 15 | | Descriptor: | (2P,3P,8S)-2-(5-fluoropyridin-2-yl)-6,6-dimethyl-3-(1H-pyrazolo[3,4-b]pyridin-4-yl)-6,7-dihydro-4H-pyrazolo[5,1-c][1,4]oxazine, Casein kinase I isoform delta | | Authors: | Thompson, A.A, Milligan, C.M, Sharma, S. | | Deposit date: | 2024-02-04 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure-Based Optimization of Selective and Brain Penetrant CK1 delta Inhibitors for the Treatment of Circadian Disruptions.
Acs Med.Chem.Lett., 15, 2024
|
|
8W1L
 
 | |
5MEV
 
 | | MCL1 FAB COMPLEX IN COMPLEX WITH COMPOUND 21 | | Descriptor: | (5~{R},13~{S},17~{S})-5-[(3,4-dichlorophenyl)methyl]-8-methyl-13-[(4-methylsulfonylphenyl)methyl]-1,4,8,12,16-pentazatricyclo[15.8.1.0^{20,25}]hexacosa-20,22,24-triene-3,7,15,26-tetrone, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Hargreaves, D. | | Deposit date: | 2016-11-16 | | Release date: | 2017-01-18 | | Last modified: | 2017-03-08 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors
Acs Med.Chem.Lett., 8, 2017
|
|
5MIJ
 
 | | X-ray structure of carboplatin-encapsulated horse spleen apoferritin | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ferritin light chain, ... | | Authors: | Pontillo, N, Ferraro, G, Helliwell, J.R, Merlino, A. | | Deposit date: | 2016-11-28 | | Release date: | 2017-03-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | X-ray Structure of the Carboplatin-Loaded Apo-Ferritin Nanocage.
ACS Med Chem Lett, 8, 2017
|
|
1RMY
 
 | | Crystal structure of AphA class B acid phosphatase/phosphotransferase ternary complex with deoxycytosine and phosphate bound to the catalytic metal | | Descriptor: | 2'-DEOXYCYTIDINE, Class B acid phosphatase, MAGNESIUM ION, ... | | Authors: | Calderone, V, Forleo, C, Benvenuti, M, Rossolini, G.M, Thaller, M.C, Mangani, S. | | Deposit date: | 2003-11-28 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights in the catalytic mechanism of AphA from Escherichia coli
To be Published
|
|
