5O1Z
 
 | |
7ACI
 
 | | In meso structure of apolipoprotein N-acyltransferase, Lnt, from Escherichia coli in 9.8 monoacylglycerol | | Descriptor: | Apolipoprotein N-acyltransferase, GLYCEROL, [(2~{S})-2,3-bis(oxidanyl)propyl] heptadec-9-enoate | | Authors: | Smithers, L, van Dalsen, L, Boland, C, Caffrey, M. | | Deposit date: | 2020-09-10 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 9.8 MAG. A new host lipid for in meso (lipid cubic phase) crystallization of integral membrane proteins
Cryst.Growth Des., 2020
|
|
6SQQ
 
 | |
6SRD
 
 | |
5NXS
 
 | |
6SQW
 
 | | Mouse dCTPase in complex with 5-Me-dCMP | | Descriptor: | 5-METHYL-2'-DEOXY-CYTIDINE-5'-MONOPHOSPHATE, MAGNESIUM ION, dCTP pyrophosphatase 1 | | Authors: | Scaletti, E.R, Claesson, M, Helleday, H, Jemth, A.S, Stenmark, P. | | Deposit date: | 2019-09-04 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The First Structure of an Active Mammalian dCTPase and its Complexes With Substrate Analogs and Products.
J.Mol.Biol., 432, 2020
|
|
5NYN
 
 | | Crystal structure of the atypical poplar thioredoxin-like2.1 in complex with gluathione | | Descriptor: | GLUTATHIONE, SULFATE ION, Thioredoxin-like protein 2.1 | | Authors: | Chibani, K, Saul, F.A, Haouz, A, Rouhier, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-02-28 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural snapshots along the reaction mechanism of the atypical poplar thioredoxin-like2.1.
FEBS Lett., 592, 2018
|
|
6ZR7
 
 | | X-ray structure of human Dscam Ig7-Ig9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Down syndrome cell adhesion molecule, ... | | Authors: | Kozak, S, Bento, I, Meijers, R. | | Deposit date: | 2020-07-11 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Homogeneously N-glycosylated proteins derived from the GlycoDelete HEK293 cell line enable diffraction-quality crystallogenesis.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6SRT
 
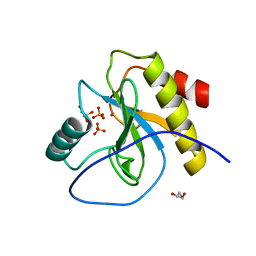 | | Endolysine N-acetylmuramoyl-L-alanine amidase LysCS from Clostridium intestinale URNW | | Descriptor: | GLYCEROL, N-acetylmuramoyl-L-alanine amidase, PHOSPHATE ION, ... | | Authors: | Hakansson, M, Al-Karadaghi, S, Plotka, M, Kaczorowska, A.-K, Kaczorowski, T. | | Deposit date: | 2019-09-06 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structure and function of endolysines LysCS, LysC from Clostridium intestinale
To Be Published
|
|
6ZLR
 
 | |
6STI
 
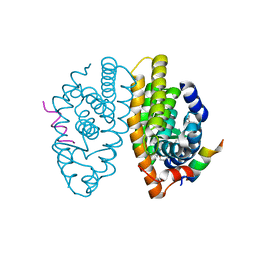 | | Crystal structure of RXRalpha LBD in complex with LG 100754 and a coactivator peptide | | Descriptor: | (2E,4E,6Z)-3-methyl-7-(5,5,8,8-tetramethyl-3-propoxy-5,6,7,8-tetrahydronaphthalen-2-yl)octa-2,4,6-trienoic acid, ACETATE ION, Nuclear receptor coactivator 2, ... | | Authors: | le Maire, A, Teyssier, C, Germain, P, Bourguet, W. | | Deposit date: | 2019-09-10 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Regulation of RXR-RAR Heterodimers by RXR- and RAR-Specific Ligands and Their Combinations.
Cells, 8, 2019
|
|
5NK9
 
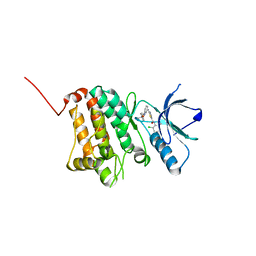 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 2e | | Descriptor: | (2~{Z})-~{N}-(2-chloranyl-6-methyl-phenyl)-2-[3-[(4-methyl-4-oxidanyl-cyclohexyl)carbamoyl]phenyl]imino-1,3-thiazolidine-5-carboxamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.588 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
6ZN8
 
 | | Crystal structure of the H. influenzae VapXD toxin-antitoxin complex | | Descriptor: | Endoribonuclease VapD, VapX | | Authors: | Bertelsen, M.B, Senissar, M, Nielsen, M.H, Bisiak, F, Cunha, M.V, Molinaro, A.L, Daines, D.A, Brodersen, D.E. | | Deposit date: | 2020-07-06 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.211 Å) | | Cite: | Structural Basis for Toxin Inhibition in the VapXD Toxin-Antitoxin System.
Structure, 29, 2021
|
|
6SUB
 
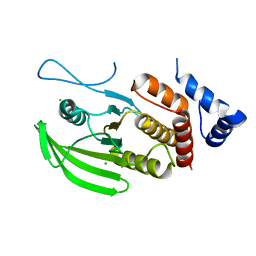 | | Human PTPRU D1 domain, reduced form | | Descriptor: | CHLORIDE ION, Receptor-type tyrosine-protein phosphatase U | | Authors: | Hay, I.M, Fearnley, G.W, Sharpe, H.J, Deane, J.E. | | Deposit date: | 2019-09-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The receptor PTPRU is a redox sensitive pseudophosphatase.
Nat Commun, 11, 2020
|
|
6SU4
 
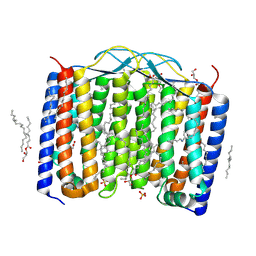 | | Crystal structure of the 48C12 heliorhodopsin in the blue form at pH 4.3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 48C12 heliorhodopsin, ACETATE ION, ... | | Authors: | Kovalev, K, Volkov, D, Astashkin, R, Alekseev, A, Gushchin, I, Gordeliy, V. | | Deposit date: | 2019-09-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution structural insights into the heliorhodopsin family.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5NKE
 
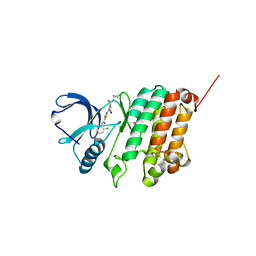 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 3a | | Descriptor: | 2-[[3-bromanyl-5-(piperidin-4-ylcarbamoyl)phenyl]amino]-~{N}-(2-chloranyl-6-methyl-phenyl)-1,3-thiazole-5-carboxamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5NKI
 
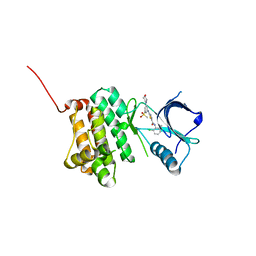 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 4b | | Descriptor: | Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[(3-methylsulfonyl-5-morpholin-4-yl-phenyl)amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.675 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
6ZNZ
 
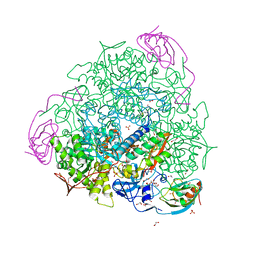 | | 1.89 A resolution 4-methylcatechol (4-methylbenzene-1,2-diol) inhibited Sporosarcina pasteurii urease | | Descriptor: | 1,2-ETHANEDIOL, HYDROXIDE ION, NICKEL (II) ION, ... | | Authors: | Mazzei, L, Cianci, M, Musiani, F, Ciurli, S. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Inhibition of Urease, a Ni-Enzyme: The Reactivity of a Key Thiol With Mono- and Di-Substituted Catechols Elucidated by Kinetic, Structural, and Theoretical Studies.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6SUT
 
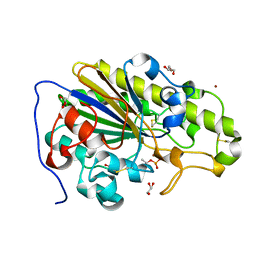 | | Crystal structure of phosphothreonine MCR-2 | | Descriptor: | BROMIDE ION, GLYCEROL, Putative integral membrane protein, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2019-09-16 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Resistance to the "last resort" antibiotic colistin: a single-zinc mechanism for phosphointermediate formation in MCR enzymes.
Chem.Commun.(Camb.), 56, 2020
|
|
5NKQ
 
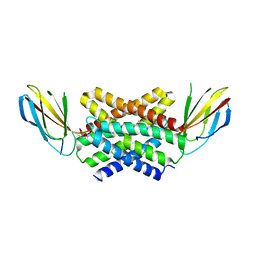 | | Crystal structure of a dual topology fluoride ion channel. | | Descriptor: | FLUORIDE ION, Monobody, Putative fluoride ion transporter CrcB, ... | | Authors: | Stockbridge, R, Miller, C, Newstead, S. | | Deposit date: | 2017-03-31 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structures of a double-barrelled fluoride ion channel.
Nature, 525, 2015
|
|
6ZU3
 
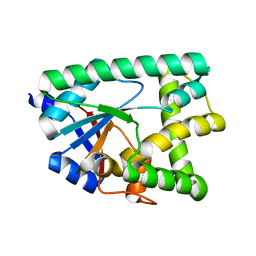 | |
6SUH
 
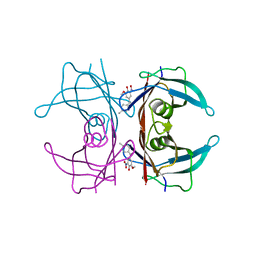 | | Crystal structure of human transthyretin in complex with 3-O-methyltolcapone, a tolcapone analogue | | Descriptor: | 3-O-methyltolcapone, Transthyretin | | Authors: | Loconte, V, Cianci, M, Menozzi, I, Sbravati, D, Sansone, F, Casnati, A, Berni, R. | | Deposit date: | 2019-09-14 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Interactions of tolcapone analogues as stabilizers of the amyloidogenic protein transthyretin.
Bioorg.Chem., 103, 2020
|
|
5NM8
 
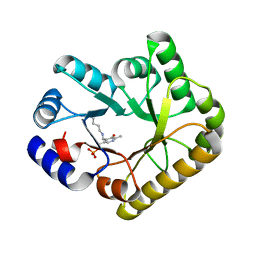 | | Structure of PipY, the COG0325 family member of Synechococcus elongatus PCC7942, with PLP bound | | Descriptor: | CALCIUM ION, PYRIDOXAL-5'-PHOSPHATE, PipY | | Authors: | Tremino, L, Forcada-Nadal, A, Contreras, A, Rubio, V. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Studies on cyanobacterial protein PipY shed light on structure, potential functions, and vitamin B6 -dependent epilepsy.
FEBS Lett., 591, 2017
|
|
6ZO3
 
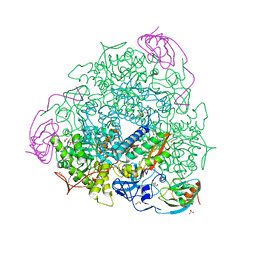 | | 1.55 A resolution 3,6-dimethylcatechol (3,6-dimethylbenzene-1,2-diol) inhibited Sporosarcina pasteurii urease | | Descriptor: | 1,2-ETHANEDIOL, HYDROXIDE ION, NICKEL (II) ION, ... | | Authors: | Mazzei, L, Cianci, M, Musiani, F, Ciurli, S. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Inhibition of Urease, a Ni-Enzyme: The Reactivity of a Key Thiol With Mono- and Di-Substituted Catechols Elucidated by Kinetic, Structural, and Theoretical Studies.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5NL0
 
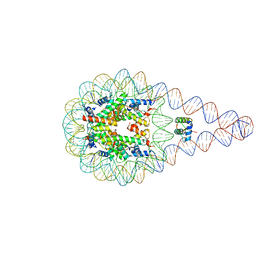 | | Crystal structure of a 197-bp palindromic 601L nucleosome in complex with linker histone H1 | | Descriptor: | DNA (197-MER), Histone H1.0-B, Histone H2A type 1, ... | | Authors: | Garcia-Saez, I, Petosa, C, Dimitrov, S. | | Deposit date: | 2017-04-03 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (5.4 Å) | | Cite: | Structure and Dynamics of a 197 bp Nucleosome in Complex with Linker Histone H1.
Mol. Cell, 66, 2017
|
|
