1Y4T
 
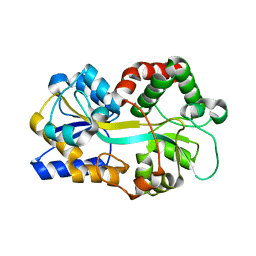 | | Ferric binding protein from Campylobacter jejuni | | Descriptor: | FE (III) ION, putative iron-uptake ABC transport system periplasmic iron-binding protein | | Authors: | Tom-Yew, S.A.L, Cui, D.T, Bekker, E.G, Murphy, M.E.P. | | Deposit date: | 2004-12-01 | | Release date: | 2005-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Anion-independent iron coordination by the Campylobacter jejuni ferric binding protein
J.Biol.Chem., 280, 2005
|
|
1Y4U
 
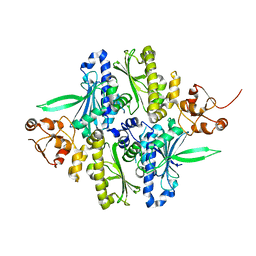 | | Conformation rearrangement of heat shock protein 90 upon ADP binding | | Descriptor: | Chaperone protein htpG | | Authors: | Huai, Q, Wang, H, Liu, Y, Kim, H, Toft, D, Ke, H. | | Deposit date: | 2004-12-01 | | Release date: | 2005-04-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the N-terminal and middle domains of E. coli Hsp90 and conformation changes upon ADP binding.
Structure, 13, 2005
|
|
1Y4V
 
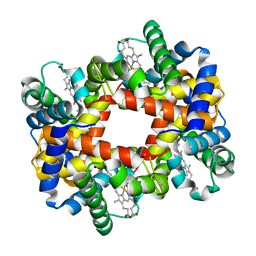 | |
1Y4W
 
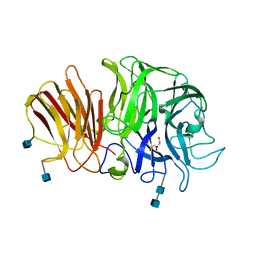 | | Crystal structure of exo-inulinase from Aspergillus awamori in spacegroup P21 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Nagem, R.A.P, Rojas, A.L, Golubev, A.M, Korneeva, O.S, Eneyskaya, E.V, Kulminskaya, A.A, Neustroev, K.N, Polikarpov, I. | | Deposit date: | 2004-12-01 | | Release date: | 2004-12-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of exo-inulinase from Aspergillus awamori: the enzyme fold and structural determinants of substrate recognition
J.Mol.Biol., 344, 2004
|
|
1Y4Y
 
 | | X-ray crystal structure of Bacillus stearothermophilus Histidine phosphocarrier protein (Hpr) | | Descriptor: | Phosphocarrier protein HPr, SULFATE ION | | Authors: | Sridharan, S, Razvi, A, Scholtz, J.M, Sacchettini, J.C. | | Deposit date: | 2004-12-01 | | Release date: | 2005-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The HPr proteins from the thermophile Bacillus stearothermophilus can form domain-swapped dimers.
J.Mol.Biol., 346, 2005
|
|
1Y4Z
 
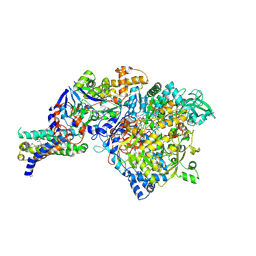 | | The crystal structure of Nitrate Reductase A, NarGHI, in complex with the Q-site inhibitor pentachlorophenol | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Boroumand, N, Palak, M, Blasco, F, Ginet, N, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2004-12-01 | | Release date: | 2005-03-08 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biochemical Characterization of a Quinol Binding Site of Escherichia coli Nitrate Reductase A
J.Biol.Chem., 280, 2005
|
|
1Y50
 
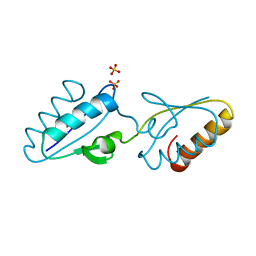 | | X-ray crystal structure of Bacillus stearothermophilus Histidine phosphocarrier protein (Hpr) F29W mutant domain_swapped dimer | | Descriptor: | Phosphocarrier protein HPr, SULFATE ION | | Authors: | Sridharan, S, Razvi, A, Scholtz, J.M, Sacchettini, J.C. | | Deposit date: | 2004-12-01 | | Release date: | 2005-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The HPr proteins from the thermophile Bacillus stearothermophilus can form domain-swapped dimers.
J.Mol.Biol., 346, 2005
|
|
1Y51
 
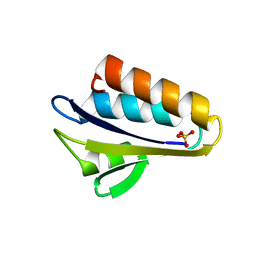 | | X-ray crystal structure of Bacillus stearothermophilus Histidine phosphocarrier protein (Hpr) F29W mutant | | Descriptor: | Phosphocarrier protein HPr, SULFATE ION | | Authors: | Sridharan, S, Razvi, A, Scholtz, J.M, Sacchettini, J.C. | | Deposit date: | 2004-12-01 | | Release date: | 2005-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The HPr proteins from the thermophile Bacillus stearothermophilus can form domain-swapped dimers.
J.Mol.Biol., 346, 2005
|
|
1Y52
 
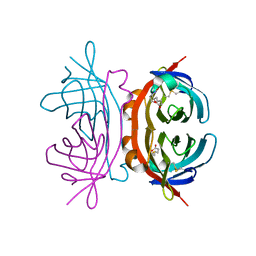 | | structure of insect cell (Baculovirus) expressed AVR4 (C122S)-biotin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Avidin-related protein 4/5, BIOTIN | | Authors: | Eisenberg-Domovich, Y, Hytonen, V.P, Wilchek, M, Bayer, E.A, Kulomaa, M.S, Livnah, O. | | Deposit date: | 2004-12-02 | | Release date: | 2005-05-24 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution crystal structure of an avidin-related protein: insight into high-affinity biotin binding and protein stability.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1Y53
 
 | | Crystal structure of bacterial expressed avidin related protein 4 (AVR4) C122S | | Descriptor: | Avidin-related protein 4/5, FORMIC ACID | | Authors: | Eisenberg-Domovich, Y, Hytonen, V.P, Wilchek, M, Bayer, E.A, Kulomaa, M.S, Livnah, O. | | Deposit date: | 2004-12-02 | | Release date: | 2005-05-24 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution crystal structure of an avidin-related protein: insight into high-affinity biotin binding and protein stability.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1Y54
 
 | | Crystal structure of the native class C beta-lactamase from Enterobacter cloacae 908R complexed with BRL42715 | | Descriptor: | (7R)-6-FORMYL-7-(1-METHYL-1H-1,2,3-TRIAZOL-4-YL)-4,7-DIHYDRO-1,4-THIAZEPINE-3-CARBOXYLIC ACID, Beta-lactamase | | Authors: | Michaux, C, Charlier, P, Frere, J.-M, Wouters, J. | | Deposit date: | 2004-12-02 | | Release date: | 2005-03-29 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of BRL 42715, C6-(N1-Methyl-1,2,3-triazolylmethylene)penem, in Complex with Enterobactercloacae 908R beta-Lactamase: Evidence for a Stereoselective Mechanism from Docking Studies
J.Am.Chem.Soc., 127, 2005
|
|
1Y55
 
 | | Crystal structure of the C122S mutant of E. Coli expressed avidin related protein 4 (AVR4)-biotin complex | | Descriptor: | Avidin-related protein 4/5, BIOTIN, FORMIC ACID | | Authors: | Eisenberg-Domovich, Y, Hytonen, V.P, Wilchek, M, Bayer, E.A, Kulomaa, M.S, Livnah, O. | | Deposit date: | 2004-12-02 | | Release date: | 2005-05-24 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution crystal structure of an avidin-related protein: insight into high-affinity biotin binding and protein stability.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1Y56
 
 | | Crystal structure of L-proline dehydrogenase from P.horikoshii | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Tsuge, H, Kawakami, R, Sakuraba, H, Ago, H, Miyano, M, Aki, K, Katunuma, N, Ohshima, T. | | Deposit date: | 2004-12-02 | | Release date: | 2005-07-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Crystal structure of a novel FAD-, FMN-, and ATP-containing L-proline dehydrogenase complex from Pyrococcus horikoshii
J.Biol.Chem., 280, 2005
|
|
1Y57
 
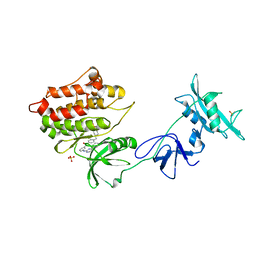 | | Structure of unphosphorylated c-Src in complex with an inhibitor | | Descriptor: | 4-[(4-METHYLPIPERAZIN-1-YL)METHYL]-N-{3-[(4-PYRIDIN-3-YLPYRIMIDIN-2-YL)AMINO]PHENYL}BENZAMIDE, Proto-oncogene tyrosine-protein kinase Src, SULFATE ION | | Authors: | Cowan-Jacob, S.W, Fendrich, G, Manley, P.W, Jahnke, W, Fabbro, D, Liebetanz, J, Meyer, T. | | Deposit date: | 2004-12-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The Crystal Structure of a c-Src Complex in an Active Conformation Suggests Possible Steps in c-Src Activation
Structure, 13, 2005
|
|
1Y58
 
 | |
1Y59
 
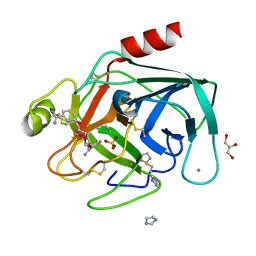 | | Dianhydrosugar-based benzamidine, factor Xa specific inhibitor in complex with bovine trypsin mutant | | Descriptor: | 2,5-BIS-O-{3-[AMINO(IMINO)METHYL]PHENYL}-1,4:3,6-DIANHYDRO-D-GLUCITOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Di Fenza, A, Heine, A, Klebe, G. | | Deposit date: | 2004-12-02 | | Release date: | 2005-12-13 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Understanding binding selectivity toward trypsin and factor Xa: the role of aromatic interactions
Chemmedchem, 2, 2007
|
|
1Y5A
 
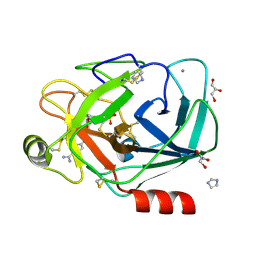 | | Dianhydrosugar-based benzamidine, factor Xa specific inhibitor in complex with bovine trypsin mutant | | Descriptor: | 2-O-{4-[AMINO(IMINO)METHYL]PHENYL}-5-O-{3-[AMINO(IMINO)METHYL]PHENYL}-1,4:3,6-DIANHYDRO-D-GLUCITOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Di Fenza, A, Heine, A, Klebe, G. | | Deposit date: | 2004-12-02 | | Release date: | 2005-12-13 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Understanding binding selectivity toward trypsin and factor Xa: the role of aromatic interactions
Chemmedchem, 2, 2007
|
|
1Y5B
 
 | | Dianhydrosugar-based benzamidine, factor Xa specific inhibitor in complex with bovine trypsin mutant | | Descriptor: | 2,5-BIS-O-{4-[AMINO(IMINO)METHYL]PHENYL}-1,4:3,6-DIANHYDRO-D-GLUCITOL, CALCIUM ION, IMIDAZOLE, ... | | Authors: | Di Fenza, A, Heine, A, Klebe, G. | | Deposit date: | 2004-12-02 | | Release date: | 2005-12-13 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Understanding binding selectivity toward trypsin and factor Xa: the role of aromatic interactions
Chemmedchem, 2, 2007
|
|
1Y5C
 
 | |
1Y5E
 
 | | Crystal structure of Molybdenum cofactor biosynthesis protein B | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, IMIDAZOLE, Molybdenum cofactor biosynthesis protein B | | Authors: | Chang, C, Zhou, M, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-02 | | Release date: | 2005-01-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Molybdenum cofactor biosynthesis protein B
TO BE PUBLISHED
|
|
1Y5F
 
 | |
1Y5H
 
 | |
1Y5I
 
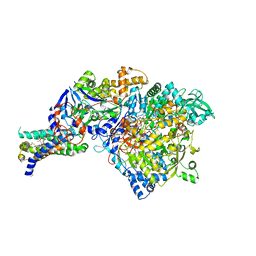 | | The crystal structure of the NarGHI mutant NarI-K86A | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, FE3-S4 CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Boroumand, N, Palak, M, Blasco, F, Ginet, N, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2004-12-02 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Biochemical Characterization of a Quinol Binding Site of Escherichia coli Nitrate Reductase A
J.Biol.Chem., 280, 2005
|
|
1Y5J
 
 | |
1Y5K
 
 | |
