8BD7
 
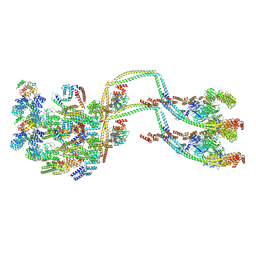 | | IFTB1 subcomplex of anterograde Intraflagellar transport trains (Chlamydomonas reinhardtii) | | Descriptor: | Clusterin-associated protein 1, IFT54, IFT70, ... | | Authors: | Lacey, S.E, Foster, H.E, Pigino, G. | | Deposit date: | 2022-10-18 | | Release date: | 2023-01-11 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (9.9 Å) | | Cite: | The molecular structure of IFT-A and IFT-B in anterograde intraflagellar transport trains.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2EF9
 
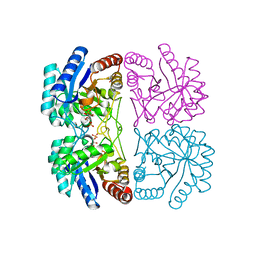 | | Structural and mechanistic changes along an engineered path from metallo to non-metallo KDO8P synthase | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, ARABINOSE-5-PHOSPHATE, PHOSPHOENOLPYRUVATE | | Authors: | Kona, F, Xu, X, Martin, P, Kuzmic, P, Gatti, D.L. | | Deposit date: | 2007-02-22 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Changes along an Engineered Path from Metallo to Nonmetallo 3-Deoxy-d-manno-octulosonate 8-Phosphate Synthases
Biochemistry, 46, 2007
|
|
2DY7
 
 | |
2EC7
 
 | | Solution Structure of Human Immunodificiency Virus Type-2 Nucleocapsid Protein | | Descriptor: | Gag polyprotein (Pr55Gag), ZINC ION | | Authors: | Matsui, T, Kodera, Y, Tanaka, T, Endoh, H, Tanaka, H, Miyauchi, E, Komatsu, H, Kohno, T, Maeda, T. | | Deposit date: | 2007-02-10 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The RNA recognition mechanism of human immunodeficiency virus (HIV) type 2 NCp8 is different from that of HIV-1 NCp7
Biochemistry, 48, 2009
|
|
4RH3
 
 | |
4RXH
 
 | |
4RXS
 
 | | The structure of GTP-dTTP-bound SAMHD1 | | Descriptor: | Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zhu, C.F, Wei, W, Peng, X, Dong, Y.H, Gong, Y, Yu, X.F. | | Deposit date: | 2014-12-11 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The mechanism of substrate-controlled allosteric regulation of SAMHD1 activated by GTP.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4RXR
 
 | | The structure of GTP-dCTP-bound SAMHD1 | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhu, C.F, Wei, W, Peng, X, Dong, Y.H, Gong, Y, Yu, X.F. | | Deposit date: | 2014-12-11 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.117 Å) | | Cite: | The mechanism of substrate-controlled allosteric regulation of SAMHD1 activated by GTP.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4RXO
 
 | | The structure of GTP-bound SAMHD1 | | Descriptor: | Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Zhu, C.F, Wei, W, Peng, X, Dong, Y.H, Gong, Y, Yu, X.F. | | Deposit date: | 2014-12-11 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The mechanism of substrate-controlled allosteric regulation of SAMHD1 activated by GTP.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8BZ1
 
 | | RNA polymerase II core pre-initiation complex with the proximal +1 nucleosome (cPIC-Nuc10W) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Abril-Garrido, J, Dienemann, C, Grabbe, F, Velychko, T, Lidschreiber, M, Wang, H, Cramer, P. | | Deposit date: | 2022-12-14 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of transcription reduction by a promoter-proximal +1 nucleosome.
Mol.Cell, 83, 2023
|
|
4RXQ
 
 | | The structure of GTP-dUTP-bound SAMHD1 | | Descriptor: | DEOXYURIDINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhu, C.F, Wei, W, Peng, X, Dong, Y.H, Gong, Y, Yu, X.F. | | Deposit date: | 2014-12-11 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The mechanism of substrate-controlled allosteric regulation of SAMHD1 activated by GTP.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4RXP
 
 | | The structure of GTP-dATP-bound SAMHD1 | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhu, C.F, Wei, W, Peng, X, Dong, Y.H, Gong, Y, Yu, X.F. | | Deposit date: | 2014-12-11 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The mechanism of substrate-controlled allosteric regulation of SAMHD1 activated by GTP.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8V5C
 
 | | IpaD (122-321) Bound to Deoxycholate | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, GLYCEROL, Invasin IpaD | | Authors: | Barker, S.A, Dickenson, N.E, Johnson, S.J. | | Deposit date: | 2023-11-30 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and functional characterization of the IpaD pi-helix reveals critical roles in DOC interaction, T3SS apparatus maturation, and Shigella virulence.
J.Biol.Chem., 300, 2024
|
|
8V7S
 
 | | IpaD (122-321) Apo Structure | | Descriptor: | Invasin IpaD | | Authors: | Barker, S.A, Dickenson, N.E, Johnson, S.J, Morales, Y. | | Deposit date: | 2023-12-04 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.708 Å) | | Cite: | Structural and functional characterization of the IpaD pi-helix reveals critical roles in DOC interaction, T3SS apparatus maturation, and Shigella virulence.
J.Biol.Chem., 300, 2024
|
|
8V7Q
 
 | |
8CZK
 
 | |
8CZL
 
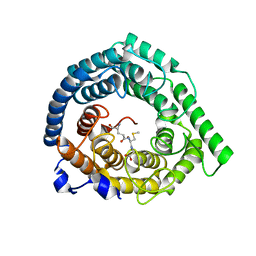 | | Human LanCL1 bound to methyl glutathione (MeGSH) | | Descriptor: | Glutathione S-transferase LANCL1, L-GAMMA-GLUTAMYL-S-METHYLCYSTEINYLGLYCINE, ZINC ION | | Authors: | Ongpipattanakul, C, Nair, S.K. | | Deposit date: | 2022-05-24 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The mechanism of thia-Michael addition catalyzed by LanC enzymes.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4UMJ
 
 | | Native structure of Farnesyl Pyrophosphate Synthase from Pseudomonas aeruginosa PA01, with bound ibandronic acid molecules. | | Descriptor: | GERANYLTRANSTRANSFERASE, IBANDRONATE, MAGNESIUM ION | | Authors: | Schmidberger, J.W, Schnell, R, Schneider, G. | | Deposit date: | 2014-05-18 | | Release date: | 2015-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Characterization of Substrate and Inhibitor Binding to Farnesyl Pyrophosphate Synthase from Pseudomonas Aeruginosa.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8D19
 
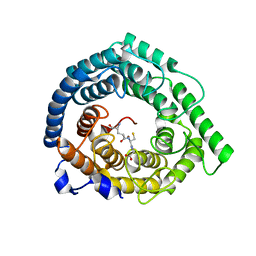 | | Human LanCL1 bound to GSH | | Descriptor: | GLUTATHIONE, Glutathione S-transferase LANCL1, ZINC ION | | Authors: | Ongpipattanakul, C, Nair, S.K. | | Deposit date: | 2022-05-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The mechanism of thia-Michael addition catalyzed by LanC enzymes.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8D0V
 
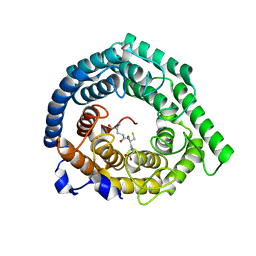 | | Human LanCL1 C264A mutant bound to GSH | | Descriptor: | GLUTATHIONE, Glutathione S-transferase LANCL1, ZINC ION | | Authors: | Ongpipattanakul, C, Nair, S.K. | | Deposit date: | 2022-05-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The mechanism of thia-Michael addition catalyzed by LanC enzymes.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5M3C
 
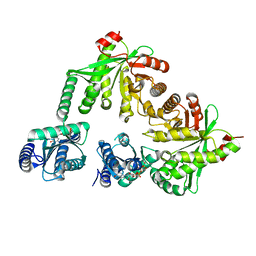 | | Structure of the hybrid domain (GGDEF-EAL) of PA0575 from Pseudomonas aeruginosa PAO1 at 2.8 Ang. with GTP and Ca2+ bound to the active site of the GGDEF domain | | Descriptor: | CALCIUM ION, Diguanylate cyclase, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Giardina, G, Brunotti, P, Cutruzzola, F, Rinaldo, S. | | Deposit date: | 2016-10-14 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into the GTP-dependent allosteric control of c-di-GMP hydrolysis from the crystal structure of PA0575 protein from Pseudomonas aeruginosa.
FEBS J., 285, 2018
|
|
6ULV
 
 | | BRD4-BD1 in complex with the cyclic peptide 4.2_1 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bromodomain-containing protein 4, Cyclic peptide 4.2_3, ... | | Authors: | Patel, K, Walshe, J.L, Walport, L.J, Mackay, J.P. | | Deposit date: | 2019-10-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cyclic peptides can engage a single binding pocket through highly divergent modes.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8J6T
 
 | | Cryo-EM structure of the double CAF-1 bound right-handed Di-tetrasome | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
8J6S
 
 | | Cryo-EM structure of the single CAF-1 bound right-handed Di-tetrasome | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
8IQF
 
 | | Cryo-EM structure of the dimeric human CAF1-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
