2VA1
 
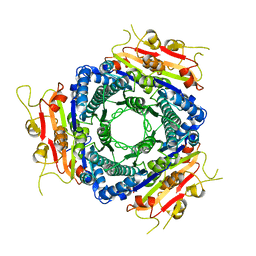 | | Crystal structure of UMP kinase from Ureaplasma parvum | | Descriptor: | PHOSPHATE ION, URIDYLATE KINASE | | Authors: | Egeblad-Welin, L, Welin, M, Wang, L, Eriksson, S. | | Deposit date: | 2007-08-28 | | Release date: | 2007-09-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Investigations of Ureaplasma Parvum Ump Kinase - a Potential Antibacterial Drug Target
FEBS J., 274, 2007
|
|
5AVM
 
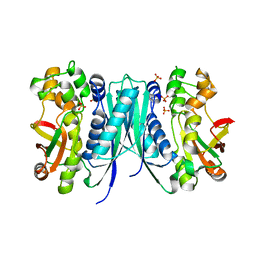 | | Crystal structures of 5-aminoimidazole ribonucleotide (AIR) synthetase, PurM, from Thermus thermophilus | | Descriptor: | Phosphoribosylformylglycinamidine cyclo-ligase, SULFATE ION | | Authors: | Kanagawa, M, Baba, S, Watanabe, Y, Nakagawa, N, Ebihara, A, Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2015-06-23 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and ligand binding of PurM proteins from Thermus thermophilus and Geobacillus kaustophilus
J.Biochem., 159, 2016
|
|
2VRH
 
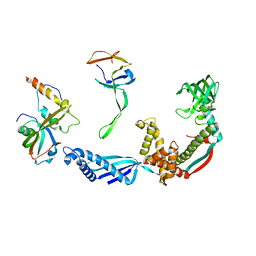 | | Structure of the E. coli trigger factor bound to a translating ribosome | | Descriptor: | 50S RIBOSOMAL PROTEIN L23, 50S RIBOSOMAL PROTEIN L24, 50S RIBOSOMAL PROTEIN L29, ... | | Authors: | Merz, F, Boehringer, D, Schaffitzel, C, Preissler, S, Hoffmann, A, Maier, T, Rutkowska, A, Lozza, J, Ban, N, Bukau, B, Deuerling, E. | | Deposit date: | 2008-04-07 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (19 Å) | | Cite: | Molecular Mechanism and Structure of Trigger Factor Bound to the Translating Ribosome.
Embo J., 27, 2008
|
|
7DW7
 
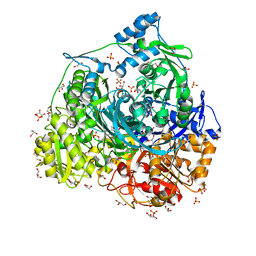 | | Crystal Structure of N1051A mutant of Formylglycinamidine Synthetase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Sharma, N, Tanwar, A.S, Anand, R. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of Coordinated Gating and Signal Transduction in Purine Biosynthetic Enzyme Formylglycinamidine Synthetase.
Acs Catalysis, 12, 2022
|
|
2YRW
 
 | | Crystal structure of GAR synthetase from Geobacillus kaustophilus | | Descriptor: | PHOSPHATE ION, Phosphoribosylglycinamide synthetase | | Authors: | Baba, S, Kanagawa, M, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria
J.Biochem., 148, 2010
|
|
2YS6
 
 | | Crystal structure of GAR synthetase from Geobacillus kaustophilus | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCINE, Phosphoribosylglycinamide synthetase | | Authors: | Baba, S, Kanagawa, M, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria
J.Biochem., 148, 2010
|
|
2YS7
 
 | | Crystal structure of GAR synthetase from Geobacillus kaustophilus | | Descriptor: | Phosphoribosylglycinamide synthetase | | Authors: | Baba, S, Kanagawa, M, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria
J.Biochem., 148, 2010
|
|
2Z02
 
 | | Crystal structure of phosphoribosylaminoimidazolesuccinocarboxamide synthase wit ATP from Methanocaldococcus jannaschii | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CITRIC ACID, Phosphoribosylaminoimidazole-succinocarboxamide synthase, ... | | Authors: | Kanagawa, M, Baba, S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-06 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of phosphoribosylaminoimidazolesuccinocarboxamide synthase from Methanocaldococcus jannaschii
To be Published
|
|
2YWR
 
 | | Crystal structure of GAR transformylase from Aquifex aeolicus | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, Phosphoribosylglycinamide formyltransferase | | Authors: | Kanagawa, M, Baba, S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-23 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structures and reaction mechanisms of the two related enzymes, PurN and PurU.
J.Biochem., 154, 2013
|
|
8FOZ
 
 | | Human IMPDH2 mutant - L245P, treated with ATP, IMP, and NAD+; filament assembly interface reconstruction | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | O'Neill, A.G, Kollman, J.M. | | Deposit date: | 2023-01-03 | | Release date: | 2023-04-19 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Neurodevelopmental disorder mutations in the purine biosynthetic enzyme IMPDH2 disrupt its allosteric regulation.
J.Biol.Chem., 299, 2023
|
|
8FUZ
 
 | | Human IMPDH2 mutant - L245P, treated with GTP, ATP, IMP, and NAD+; filament assembly interface reconstruction | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | O'Neill, A.G, Kollman, J.M. | | Deposit date: | 2023-01-18 | | Release date: | 2023-04-19 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Neurodevelopmental disorder mutations in the purine biosynthetic enzyme IMPDH2 disrupt its allosteric regulation.
J.Biol.Chem., 299, 2023
|
|
8FUR
 
 | | Crystal structure of human IDO1 with compound 11 | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-(4-methylphenyl)-N'-[(1P,2'P)-4-propoxy-5-propyl-2'-(1H-tetrazol-5-yl)[1,1'-biphenyl]-3-yl]urea | | Authors: | Critton, D.A, Lewis, H.A. | | Deposit date: | 2023-01-18 | | Release date: | 2023-04-26 | | Last modified: | 2023-05-03 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Synthesis and biological evaluation of biaryl alkyl ethers as inhibitors of IDO1.
Bioorg.Med.Chem.Lett., 88, 2023
|
|
8FDY
 
 | | Human GAR transformylase in complex with GAR substrate and AGF132 inhibitor | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-{5-[4-(2-amino-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidin-6-yl)butyl]pyridine-2-carbonyl}-L-glutamic acid, Phosphoribosylglycinamide formyltransferase | | Authors: | Wong-Roushar, J, Dann III, C.E. | | Deposit date: | 2022-12-05 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Multitargeted 6-Substituted Thieno[2,3- d ]pyrimidines as Folate Receptor-Selective Anticancer Agents that Inhibit Cytosolic and Mitochondrial One-Carbon Metabolism.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8FJW
 
 | | Human GAR transformylase in complex with GAR substrate and AGF347 inhibitor | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-{4-[4-(2-amino-4-oxo-3,4-dihydro-5H-pyrrolo[3,2-d]pyrimidin-5-yl)butyl]-2-fluorobenzoyl}-L-glutamic acid, Trifunctional purine biosynthetic protein adenosine-3 | | Authors: | Wong-Roushar, J, Dann III, C.E. | | Deposit date: | 2022-12-20 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure-Based Design of Transport-Specific Multitargeted One-Carbon Metabolism Inhibitors in Cytosol and Mitochondria.
J.Med.Chem., 66, 2023
|
|
2DJL
 
 | | Crystal structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with succinate | | Descriptor: | COBALT HEXAMMINE(III), FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Inaoka, D.K, Shimizu, H, Sakamoto, K, Shiba, T, Kurisu, G, Nara, T, Aoki, T, Harada, S, Kita, K. | | Deposit date: | 2006-04-04 | | Release date: | 2007-06-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with succinate
To be Published
|
|
2DG9
 
 | |
2BNF
 
 | | The structure of E. coli UMP kinase in complex with UTP | | Descriptor: | GLYCEROL, URIDINE 5'-TRIPHOSPHATE, URIDYLATE KINASE | | Authors: | Briozzo, P, Evrin, C, Meyer, P, Assairi, L, Joly, N, Barzu, O, Gilles, A.M. | | Deposit date: | 2005-03-23 | | Release date: | 2005-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of Escherichia Coli Ump Kinase Differs from that of Other Nucleoside Monophosphate Kinases and Sheds New Light on Enzyme Regulation.
J.Biol.Chem., 280, 2005
|
|
8G8F
 
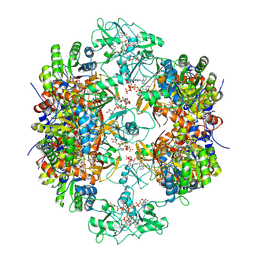 | | Human IMPDH2 mutant - L245P, treated with ATP, IMP, and NAD+; extended filament segment reconstruction | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 2, ... | | Authors: | O'Neill, A.G, Kollman, J.M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-04-19 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Neurodevelopmental disorder mutations in the purine biosynthetic enzyme IMPDH2 disrupt its allosteric regulation.
J.Biol.Chem., 299, 2023
|
|
2DG3
 
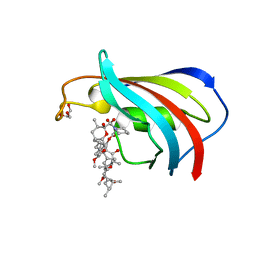 | | Wildtype FK506-binding protein complexed with Rapamycin | | Descriptor: | FK506-binding protein 1A, GLYCEROL, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Buckle, A.M. | | Deposit date: | 2006-03-08 | | Release date: | 2006-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Energetic and structural analysis of the role of tryptophan 59 in FKBP12
Biochemistry, 42, 2003
|
|
2DGB
 
 | |
8G7M
 
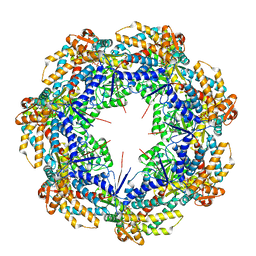 | | ATP-bound mtHsp60 V72I focus | | Descriptor: | 60 kDa heat shock protein, mitochondrial, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8G7L
 
 | | ATP-bound mtHsp60 V72I | | Descriptor: | 60 kDa heat shock protein, mitochondrial, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8G7O
 
 | | ATP- and mtHsp10-bound mtHsp60 V72I focus | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8G7N
 
 | | ATP- and mtHsp10-bound mtHsp60 V72I | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8G7J
 
 | | mtHsp60 V72I apo | | Descriptor: | 60 kDa heat shock protein, mitochondrial | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
