400D
 
 | |
6N4G
 
 | |
3QK4
 
 | |
7DTA
 
 | |
1EZN
 
 | | SOLUTION STRUCTURE OF A DNA THREE-WAY JUNCTION | | Descriptor: | DNA THREE-WAY JUNCTION | | Authors: | van Buuren, B.N.M, Overmars, F.J, Ippel, J.H, Altona, C, Wijmenga, S.S. | | Deposit date: | 2000-05-11 | | Release date: | 2001-04-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA three-way junction containing two unpaired thymidine bases. Identification of sequence features that decide conformer selection.
J.Mol.Biol., 304, 2000
|
|
1F3S
 
 | | Solution Structure of DNA Sequence GGGTTCAGG Forms GGGG Tetrade and G(C-A) Triad. | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*TP*CP*AP*GP*G)-3') | | Authors: | Kettani, A, Basu, G, Gorin, A, Majumdar, A, Skripkin, E, Patel, D.J. | | Deposit date: | 2000-06-06 | | Release date: | 2000-11-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A two-stranded template-based approach to G.(C-A) triad formation: designing novel structural elements into an existing DNA framework.
J.Mol.Biol., 301, 2000
|
|
5XJZ
 
 | | Structure of DNA1-Ag complex | | Descriptor: | DNA (5'-D(*GP*CP*AP*CP*GP*CP*GP*C)-3'), SILVER ION, SPERMINE | | Authors: | Liu, H.H, Gan, J.H. | | Deposit date: | 2017-05-04 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | A DNA Structure Containing AgI -Mediated G:G and C:C Base Pairs
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
1VTV
 
 | | Molecular structure of (M5DC-DG)3: The role of the methyl group on 5-methyl cytosine in stabilizing Z-DNA | | Descriptor: | DNA (5'-D(*(CH3)CP*GP*(CH3)CP*GP*(CH3)CP*G)-3') | | Authors: | Fujii, S, Wang, A.H.-J, Van Der Marel, G.A, Van Boom, J.H, Rich, A. | | Deposit date: | 1989-01-10 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular Structure of (m5dC-dG)3: The Role of the Methyl Group on 5-Methyl Cytosine in Stabilizing Z-DNA
Nucleic Acids Res., 10, 1982
|
|
2ZCJ
 
 | | Ternary structure of the Glu119Gln M.HhaI, C5-Cytosine DNA methyltransferase, with unmodified DNA and AdoHcy | | Descriptor: | DNA (5'-D(*DGP*DAP*DTP*DAP*DGP*DCP*DGP*DCP*DTP*DAP*DTP*DC)-3'), DNA (5'-D(*DTP*DGP*DAP*DTP*DAP*DGP*DCP*DGP*DCP*DTP*DAP*DTP*DC)-3'), Modification methylase HhaI, ... | | Authors: | Shieh, F.K, Reich, N.O. | | Deposit date: | 2007-11-09 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | AdoMet-dependent Methyl-transfer: Glu(119) Is Essential for DNA C5-Cytosine Methyltransferase M.HhaI
J.Mol.Biol., 373, 2007
|
|
5UUF
 
 | |
3MHT
 
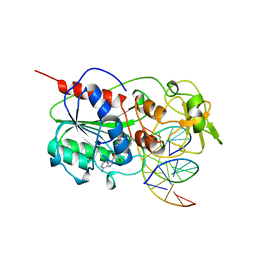 | | TERNARY STRUCTURE OF HHAI METHYLTRANSFERASE WITH UNMODIFIED DNA AND ADOHCY | | Descriptor: | DNA (5'-D(*GP*AP*TP*AP*GP*CP*GP*CP*TP*AP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*TP*AP*GP*CP*GP*CP*TP*AP*TP*C)-3'), PROTEIN (HHAI METHYLTRANSFERASE (E.C.2.1.1.73)), ... | | Authors: | O'Gara, M, Klimasauskas, S, Roberts, R.J, Cheng, X. | | Deposit date: | 1996-07-24 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Enzymatic C5-cytosine methylation of DNA: mechanistic implications of new crystal structures for HhaL methyltransferase-DNA-AdoHcy complexes.
J.Mol.Biol., 261, 1996
|
|
7FEF
 
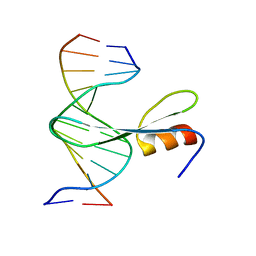 | | Crystal structure of AtMBD6 with DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain-containing protein 6 | | Authors: | Wu, Z.B, Liu, K, Min, J.R. | | Deposit date: | 2021-07-19 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Family-wide Characterization of Methylated DNA Binding Ability of Arabidopsis MBDs.
J.Mol.Biol., 434, 2022
|
|
4QEP
 
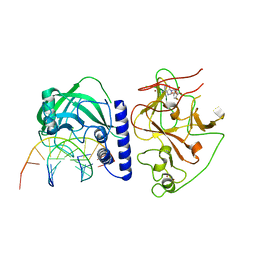 | | crystal structure of KRYPTONITE in complex with mCHG DNA and SAH | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*CP*TP*GP*AP*GP*TP*AP*CP*CP*AP*T)-3'), DNA (5'-D(*GP*GP*TP*AP*CP*TP*(5CM)P*AP*GP*CP*AP*GP*TP*AP*T)-3'), Histone-lysine N-methyltransferase, ... | | Authors: | Du, J, Li, S, Patel, D.J. | | Deposit date: | 2014-05-17 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanism of DNA Methylation-Directed Histone Methylation by KRYPTONITE.
Mol.Cell, 55, 2014
|
|
6KKV
 
 | |
4QEN
 
 | | crystal structure of KRYPTONITE in complex with mCHH DNA and SAH | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*AP*TP*GP*AP*GP*TP*AP*CP*CP*AP*T)-3'), DNA (5'-D(*GP*GP*TP*AP*CP*TP*(5CM)P*AP*TP*CP*AP*GP*TP*AP*T)-3'), Histone-lysine N-methyltransferase, ... | | Authors: | Du, J, Li, S, Patel, D.J. | | Deposit date: | 2014-05-17 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Mechanism of DNA Methylation-Directed Histone Methylation by KRYPTONITE.
Mol.Cell, 55, 2014
|
|
3PVI
 
 | |
2Z6U
 
 | |
4D05
 
 | | Structure and activity of a minimal-type ATP-dependent DNA ligase from a psychrotolerant bacterium | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP-DEPENDENT DNA LIGASE, MAGNESIUM ION, ... | | Authors: | Williamson, A, Rothweiler, U, Leiros, H.-K.S. | | Deposit date: | 2014-04-24 | | Release date: | 2014-11-12 | | Last modified: | 2019-06-26 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Enzyme-Adenylate Structure of a Bacterial ATP-Dependent DNA Ligase with a Minimized DNA-Binding Surface
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7WM3
 
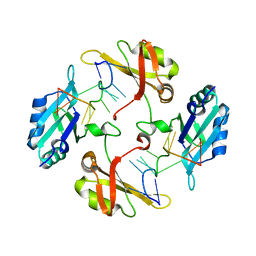 | | hnRNP A2/B1 RRMs in complex with single-stranded DNA | | Descriptor: | DNA (5'-D(P*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), Heterogeneous nuclear ribonucleoproteins A2/B1 | | Authors: | Abula, A, Liu, Y, Guo, H, Li, T, Ji, X. | | Deposit date: | 2022-01-14 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural Insight Into hnRNP A2/B1 Homodimerization and DNA Recognition.
J.Mol.Biol., 435, 2022
|
|
5UZF
 
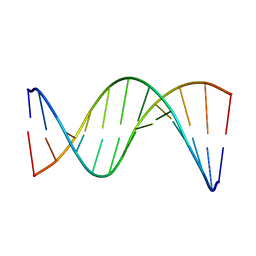 | | Insights into Watson-Crick/Hoogsteen Breathing Dynamics and Damage Repair from the Solution Structure and Dynamic Ensemble of DNA Duplexes containing m1A - A6-DNA structure | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*TP*TP*TP*TP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*AP*AP*AP*AP*AP*TP*CP*G)-3') | | Authors: | Sathyamoorthy, B, Shi, H, Xue, Y, Al-Hashimi, H.M. | | Deposit date: | 2017-02-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into Watson-Crick/Hoogsteen breathing dynamics and damage repair from the solution structure and dynamic ensemble of DNA duplexes containing m1A.
Nucleic Acids Res., 45, 2017
|
|
5UZD
 
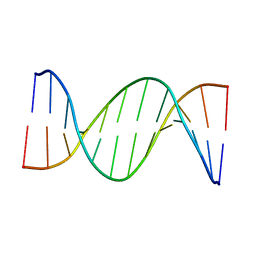 | | Insights into Watson-Crick/Hoogsteen Breathing Dynamics and Damage Repair from the Solution Structure and Dynamic Ensemble of DNA Duplexes containing m1A - A2-DNA structure | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*GP*AP*TP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*AP*TP*CP*GP*AP*TP*GP*C)-3') | | Authors: | Sathyamoorthy, B, Shi, H, Xue, Y, Al-Hashimi, H.M. | | Deposit date: | 2017-02-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into Watson-Crick/Hoogsteen breathing dynamics and damage repair from the solution structure and dynamic ensemble of DNA duplexes containing m1A.
Nucleic Acids Res., 45, 2017
|
|
3PT6
 
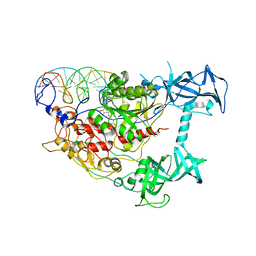 | | Crystal structure of mouse DNMT1(650-1602) in complex with DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*CP*GP*GP*AP*GP*GP*CP*TP*CP*AP*CP*GP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*CP*GP*TP*GP*AP*GP*CP*CP*TP*CP*CP*GP*CP*AP*GP*G)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation.
Science, 331, 2011
|
|
7X5K
 
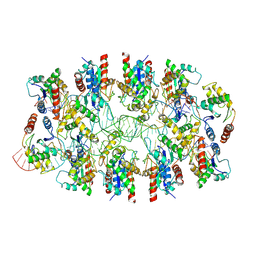 | | Tir-dsDNA complex, the initial binding state | | Descriptor: | DNA (43-MER), Flax rust resistance protein | | Authors: | Tan, Y, Xu, C, Yu, D, Song, W, Wu, B, Schulze-Lefert, P, Chai, J. | | Deposit date: | 2022-03-04 | | Release date: | 2022-06-08 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | TIR domains of plant immune receptors are 2',3'-cAMP/cGMP synthetases mediating cell death.
Cell, 185, 2022
|
|
2RPD
 
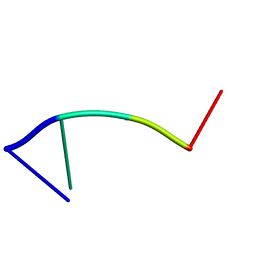 | | Mhr1p-bound ssDNA | | Descriptor: | DNA (5'-D(*DTP*DAP*DCP*DG)-3') | | Authors: | Masuda, T, Ito, Y, Shibata, T, Mikawa, T. | | Deposit date: | 2008-05-15 | | Release date: | 2009-05-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A non-canonical DNA structure enables homologous recombination in various genetic systems
J.Biol.Chem., 284, 2009
|
|
2RPF
 
 | | RecO-bound ssDNA | | Descriptor: | DNA (5'-D(*DTP*DAP*DCP*DG)-3') | | Authors: | Masuda, T, Ito, Y, Shibata, T, Mikawa, T. | | Deposit date: | 2008-05-15 | | Release date: | 2009-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A non-canonical DNA structure enables homologous recombination in various genetic systems
J.Biol.Chem., 284, 2009
|
|
