3BM1
 
 | |
3CZG
 
 | | Crystal Structure Analysis of Sucrose hydrolase (SUH)-glucose complex | | Descriptor: | Sucrose hydrolase, alpha-D-glucopyranose | | Authors: | Kim, M.I, Rhee, S. | | Deposit date: | 2008-04-29 | | Release date: | 2008-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures and mutagenesis of sucrose hydrolase from Xanthomonas axonopodis pv. glycines: insight into the exclusively hydrolytic amylosucrase fold.
J.Mol.Biol., 380, 2008
|
|
3K9P
 
 | | The crystal structure of E2-25K and ubiquitin complex | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 K | | Authors: | Kang, G.B, Ko, S, Song, S.M, Lee, W, Eom, S.H. | | Deposit date: | 2009-10-16 | | Release date: | 2010-09-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of E2-25K/UBB+1 interaction leading to proteasome inhibition and neurotoxicity
J.Biol.Chem., 285, 2010
|
|
3CZE
 
 | | Crystal Structure Analysis of Sucrose hydrolase (SUH)- Tris complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Sucrose hydrolase | | Authors: | Kim, M.I, Rhee, S. | | Deposit date: | 2008-04-29 | | Release date: | 2008-07-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures and mutagenesis of sucrose hydrolase from Xanthomonas axonopodis pv. glycines: insight into the exclusively hydrolytic amylosucrase fold.
J.Mol.Biol., 380, 2008
|
|
3JZ0
 
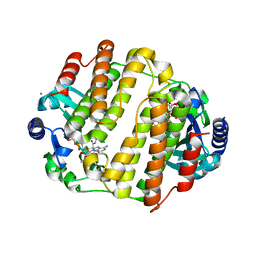 | | LinB complexed with clindamycin and AMPCPP | | Descriptor: | CLINDAMYCIN, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, Lincosamide nucleotidyltransferase, ... | | Authors: | Morar, M, Wright, G.D. | | Deposit date: | 2009-09-22 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of the lincosamide antibiotic adenylyltransferase LinB.
Structure, 17, 2009
|
|
3JRW
 
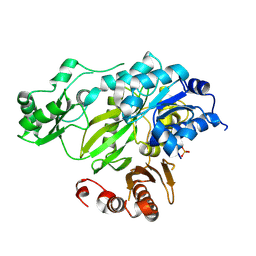 | | Phosphorylated BC domain of ACC2 | | Descriptor: | Acetyl-CoA carboxylase 2 | | Authors: | Cho, Y.S, Lee, J.I, Shin, D, Kim, H.T, Lee, T.G, Heo, Y.S. | | Deposit date: | 2009-09-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mechanism for the regulation of human ACC2 through phosphorylation by AMPK
Biochem.Biophys.Res.Commun., 391, 2010
|
|
3JRX
 
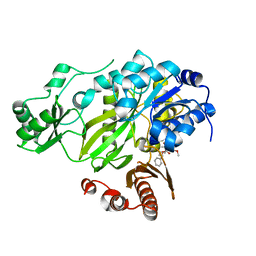 | | Crystal structure of the BC domain of ACC2 in complex with soraphen A | | Descriptor: | Acetyl-CoA carboxylase 2, SORAPHEN A | | Authors: | Cho, Y.S, Lee, J.I, Shin, D, Kim, H.T, Lee, T.G, Heo, Y.S. | | Deposit date: | 2009-09-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism for the regulation of human ACC2 through phosphorylation by AMPK.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
3JYY
 
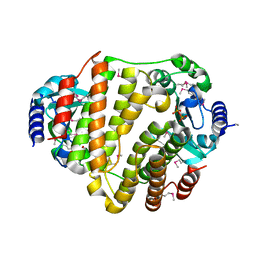 | | SeMet LinB complexed with PPi | | Descriptor: | Lincosamide nucleotidyltransferase, MAGNESIUM ION, PYROPHOSPHATE | | Authors: | Morar, M, Wright, G.D. | | Deposit date: | 2009-09-22 | | Release date: | 2009-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanism of the lincosamide antibiotic adenylyltransferase LinB.
Structure, 17, 2009
|
|
3BF8
 
 | |
3AQ3
 
 | |
3AQ2
 
 | |
3AQ4
 
 | |
3BAT
 
 | |
3MCL
 
 | |
3BAS
 
 | |
3CZL
 
 | |
3BWO
 
 | | L-tryptophan aminotransferase | | Descriptor: | L-tryptophan aminotransferase | | Authors: | Ferrer, J.-L, Noel, J.P, Pojer, F, Bowman, M, Chory, J, Tao, Y. | | Deposit date: | 2008-01-10 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for the shade avoidance response of plants
To be Published
|
|
3BL2
 
 | | Crystal Structure of M11, the BCL-2 Homolog of Murine Gamma-herpesvirus 68, Complexed with Mouse Beclin1 (residues 106-124) | | Descriptor: | Beclin-1, V-bcl-2 | | Authors: | Oh, B.-H, Woo, J.-S, Ku, B. | | Deposit date: | 2007-12-10 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Bases for the Inhibition of Autophagy and Apoptosis by Viral BCL-2 of Murine gamma-Herpesvirus 68
Plos Pathog., 4, 2008
|
|
3KJ4
 
 | | Structure of rat Nogo receptor bound to 1D9 antagonist antibody | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab fragment 1D9 heavy chain, ... | | Authors: | Silvian, L.F. | | Deposit date: | 2009-11-02 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Resolution of disulfide heterogeneity in Nogo receptor 1 fusion proteins by molecular engineering.
Biotechnol Appl Biochem, 57, 2010
|
|
3BM2
 
 | |
3BWN
 
 | | L-tryptophan aminotransferase | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, L-tryptophan aminotransferase, PHENYLALANINE, ... | | Authors: | Ferrer, J.-L, Noel, J.P, Pojer, F, Bowman, M, Chory, J, Tao, Y. | | Deposit date: | 2008-01-10 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants
Cell(Cambridge,Mass.), 133, 2008
|
|
3O11
 
 | | Anti-beta-amyloid antibody c706 fab in space group c2 | | Descriptor: | C706 HEAVY CHAIN variable region, Ig gamma-1 chain C region chimera, C706 LIGHT CHAIN variable region, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2010-07-20 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | His-tag binding by antibody C706 mimics beta-amyloid recognition.
J.Mol.Recognit., 24, 2011
|
|
7KWN
 
 | |
3SL7
 
 | | Crystal structure of CBS-pair protein, CBSX2 from Arabidopsis thaliana | | Descriptor: | ACETATE ION, CBS domain-containing protein CBSX2, GLYCEROL | | Authors: | Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2011-06-24 | | Release date: | 2011-11-09 | | Last modified: | 2013-10-09 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Single cystathionine beta-synthase domain-containing proteins modulate development by regulating the thioredoxin system in Arabidopsis
Plant Cell, 23, 2011
|
|
5OAH
 
 | | THE PERIPLASMIC BINDING PROTEIN CEUE OF CAMPYLOBACTER JEJUNI BINDS THE IRON(III) COMPLEX OF Azotochelin | | Descriptor: | Azotochelin, Enterochelin ABC transporter substrate-binding protein, FE (III) ION | | Authors: | Raines, A.D.J, Blagova, E, Dodson, E.J, Wilson, K.S, Duhme-Klair, A.K. | | Deposit date: | 2017-06-22 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Redox-switchable siderophore anchor enables reversible artificial metalloenzyme assembly
Nat Catal, 2018
|
|
