5TD5
 
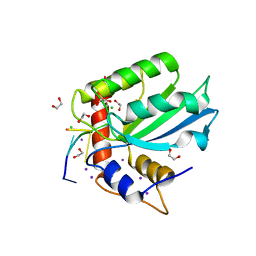 | | Crystal Structure of Human APOBEC3B variant complexed with ssDNA | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(P*TP*TP*CP*AP*T)-3'), ... | | Authors: | Shi, K, Banerjee, S, Kurahashi, K, Aihara, H. | | Deposit date: | 2016-09-16 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.718 Å) | | Cite: | Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B.
Nat. Struct. Mol. Biol., 24, 2017
|
|
8GEN
 
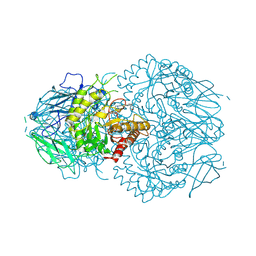 | | E. eligens beta-glucuronidase bound to UNC10201652-glucuronide | | Descriptor: | 8-(4-beta-D-glucopyranuronosylpiperazin-1-yl)-5-(morpholin-4-yl)-1,2,3,4-tetrahydro[1,2,3]triazino[4',5':4,5]thieno[2,3 -c]isoquinoline, Beta-glucuronidase | | Authors: | Simpson, J.B, Redinbo, M.R. | | Deposit date: | 2023-03-07 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Gut microbial beta-glucuronidases influence endobiotic homeostasis and are modulated by diverse therapeutics.
Cell Host Microbe, 32, 2024
|
|
5K6R
 
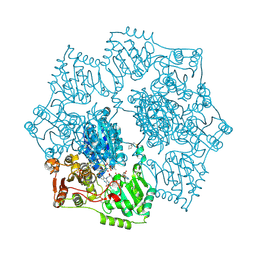 | | Crystal structure of Arabidopsis thaliana acetohydroxyacid synthase in complex with a sulfonylamino-carbonyl-triazolinone herbicide, thiencarbazone-methyl | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Acetolactate synthase, ... | | Authors: | Garcia, M.D, Lonhienne, T, Guddat, L.W. | | Deposit date: | 2016-05-25 | | Release date: | 2017-02-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.734 Å) | | Cite: | Comprehensive understanding of acetohydroxyacid synthase inhibition by different herbicide families.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5DLD
 
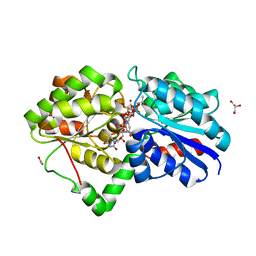 | |
3L16
 
 | | Discovery of (thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer | | Descriptor: | 5-(6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)pyridin-2-amine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M, Wiesmann, C. | | Deposit date: | 2009-12-10 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of (Thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer.
J.Med.Chem., 53, 2010
|
|
4Z4R
 
 | | Crystal structure of GII.10 P domain in complex with 300mM fucose | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, beta-L-fucopyranose | | Authors: | Koromyslova, A.D, Leuthold, M.M, Hansman, G.S. | | Deposit date: | 2015-04-02 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | The sweet quartet: Binding of fucose to the norovirus capsid.
Virology, 483, 2015
|
|
6DMK
 
 | | A multiconformer ligand model of an isoxazolyl-benzimidazole ligand bound to the bromodomain of human CREBBP | | Descriptor: | 1,2-ETHANEDIOL, 5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[2-(morpholin-4-yl)ethyl]-2-(2-phenylethyl)-1H-benzimidazole, CREB-binding protein, ... | | Authors: | Hudson, B.M, van Zundert, G, Keedy, D.A, Fonseca, R, Heliou, A, Suresh, P, Borrelli, K, Day, T, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | qFit-ligand Reveals Widespread Conformational Heterogeneity of Drug-Like Molecules in X-Ray Electron Density Maps.
J. Med. Chem., 61, 2018
|
|
5K9W
 
 | | Protein Tyrosine Phosphatase 1B (1-301) in complex with TCS401, closed state | | Descriptor: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2016-06-01 | | Release date: | 2017-03-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Conformational Rigidity and Protein Dynamics at Distinct Timescales Regulate PTP1B Activity and Allostery.
Mol. Cell, 65, 2017
|
|
2QTW
 
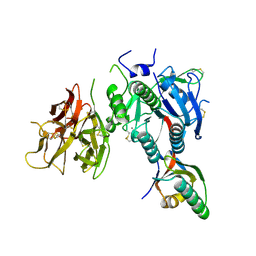 | | The Crystal Structure of PCSK9 at 1.9 Angstroms Resolution Reveals structural homology to Resistin within the C-terminal domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Proprotein convertase subtilisin/kexin type 9, ... | | Authors: | Hampton, E.N, Knuth, M.W, Li, J, Harris, J.L, Lesley, S.A, Spraggon, G. | | Deposit date: | 2007-08-02 | | Release date: | 2007-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The self-inhibited structure of full-length PCSK9 at 1.9 A reveals structural homology with resistin within the C-terminal domain.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4MCW
 
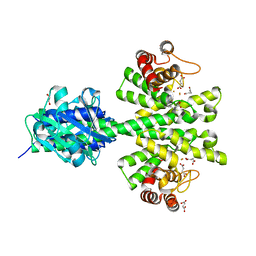 | | Crystal structure of a HD-GYP domain (a cyclic-di-GMP phosphodiesterase) containing a tri-nuclear metal centre | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, IMIDAZOLE, ... | | Authors: | Bellini, D, Walsh, M.A, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2013-08-21 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of an HD-GYP domain cyclic-di-GMP phosphodiesterase reveals an enzyme with a novel trinuclear catalytic iron centre.
Mol.Microbiol., 91, 2014
|
|
2QE7
 
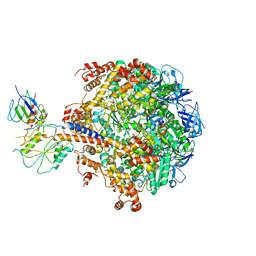 | | Crystal structure of the f1-atpase from the thermoalkaliphilic bacterium bacillus sp. ta2.a1 | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit epsilon, ... | | Authors: | Stocker, A, Keis, S, Vonck, J, Cook, G.M, Dimroth, P. | | Deposit date: | 2007-06-25 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | The Structural Basis for Unidirectional Rotation of Thermoalkaliphilic F(1)-ATPase.
Structure, 15, 2007
|
|
6JVJ
 
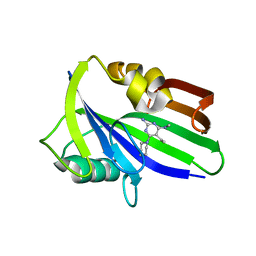 | | Crystal structure of human MTH1 in complex with compound MI1006 | | Descriptor: | 5-ethyl-N4-methyl-6-piperidin-1-yl-pyrimidine-2,4-diamine, 7,8-dihydro-8-oxoguanine triphosphatase | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6DT1
 
 | | Crystal structure of the ligase from bacteriophage T4 complexed with DNA intermediate | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2018-06-14 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | T4 DNA ligase structure reveals a prototypical ATP-dependent ligase with a unique mode of sliding clamp interaction.
Nucleic Acids Res., 46, 2018
|
|
6JVR
 
 | | Crystal structure of human MTH1 in complex with compound MI1026 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-methyl-6-piperidin-1-yl-pyrimidine-2,4-diamine | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
8OIN
 
 | | 55S mammalian mitochondrial ribosome with mtRF1 and P-site tRNA | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S15, ... | | Authors: | Saurer, M, Leibundgut, M, Scaiola, A, Schoenhut, T, Ban, N. | | Deposit date: | 2023-03-23 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis of translation termination at noncanonical stop codons in human mitochondria.
Science, 380, 2023
|
|
3DMI
 
 | | Crystallization and Structural Analysis of Cytochrome c6 from the Diatom Phaeodactylum tricornutum at 1.5 A resolution | | Descriptor: | HEME C, MAGNESIUM ION, cytochrome c6 | | Authors: | Akazaki, H, Kawai, F, Hosokawa, M, Hama, T, Hirano, T, Lim, B.-K, Sakurai, N, Hakamata, W, Park, S.-Y, Nishio, T, Oku, T. | | Deposit date: | 2008-07-01 | | Release date: | 2009-03-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallization and structural analysis of cytochrome c(6) from the diatom Phaeodactylum tricornutum at 1.5 A resolution.
Biosci.Biotechnol.Biochem., 73, 2009
|
|
8OIT
 
 | | 39S human mitochondrial large ribosomal subunit with mtRF1 and P-site tRNA | | Descriptor: | 16S rRNA, 39S ribosomal protein L1, mitochondrial, ... | | Authors: | Saurer, M, Leibundgut, M, Scaiola, A, Schoenhut, T, Ban, N. | | Deposit date: | 2023-03-23 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis of translation termination at noncanonical stop codons in human mitochondria.
Science, 380, 2023
|
|
6KIE
 
 | | Crystal structure of human leucyl-tRNA synthetase, Leu-AMS-bound form | | Descriptor: | 5'-O-(L-leucylsulfamoyl)adenosine, LEUCINE, Leucine--tRNA ligase, ... | | Authors: | Kim, S, Son, J, Kim, S, Hwang, K.Y. | | Deposit date: | 2019-07-18 | | Release date: | 2021-01-27 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Leucine-sensing mechanism of leucyl-tRNA synthetase 1 for mTORC1 activation.
Cell Rep, 35, 2021
|
|
6KIL
 
 | | N21Q mutant thioredoxin from Halobacterium salinarum NRC-1 | | Descriptor: | Thioredoxin | | Authors: | Arai, S, Shibazaki, C, Shimizu, R, Adachi, M, Ishibashi, M, Tokunaga, H, Tokunaga, M. | | Deposit date: | 2019-07-19 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Catalytic mechanism and evolutional characteristics of thioredoxin from Halobacterium salinarum NRC-1.
Acta Crystallogr.,Sect.D, 76, 2020
|
|
5ZS7
 
 | |
7WXM
 
 | |
4MM1
 
 | | GGGPS from Methanothermobacter thermautotrophicus | | Descriptor: | Geranylgeranylglyceryl phosphate synthase, SN-GLYCEROL-1-PHOSPHATE, TRIETHYLENE GLYCOL | | Authors: | Rajendran, C, Peterhoff, D, Beer, B, Kumpula, E.P, Kapetaniou, E, Guldan, H, Wierenga, R.K, Sterner, R, Babinger, P. | | Deposit date: | 2013-09-07 | | Release date: | 2014-06-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8004 Å) | | Cite: | A comprehensive analysis of the geranylgeranylglyceryl phosphate synthase enzyme family identifies novel members and reveals mechanisms of substrate specificity and quaternary structure organization.
Mol.Microbiol., 92, 2014
|
|
5TDX
 
 | | Resurrected Ancestral Hydroxynitrile Lyase from Flowering Plants | | Descriptor: | Ancestral Hydroxynitrile Lyase 1, GLYCEROL | | Authors: | Jones, B.J, Evans, R, Wilmot, C.M, Kazlauskas, R.J. | | Deposit date: | 2016-09-20 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Larger active site in an ancestral hydroxynitrile lyase increases catalytically promiscuous esterase activity.
Plos One, 15, 2020
|
|
7DHN
 
 | | The co-crystal structure of DYRK2 with a small molecule inhibitor 20 | | Descriptor: | 7-methoxy-2-methylsulfanyl-9-(piperidin-4-ylmethylsulfanyl)-[1,3]thiazolo[5,4-b]quinoline, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Wei, T, Xiao, J. | | Deposit date: | 2020-11-16 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Selective inhibition reveals the regulatory function of DYRK2 in protein synthesis and calcium entry.
Elife, 11, 2022
|
|
9BVP
 
 | | Vitamin K-dependent gamma-carboxylase with TMG2 propeptide and glutamate-rich region and with vitamin K hydroquinone | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, W, Liu, B, Cao, Q. | | Deposit date: | 2024-05-20 | | Release date: | 2025-01-22 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis of vitamin-K-driven gamma-carboxylation at the membrane interface.
Nature, 639, 2025
|
|
