8U1T
 
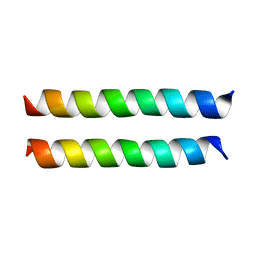 | | SARS-CoV-2 Envelope Protein Transmembrane Domain: Dimeric Structure Determined by Solid-State NMR | | Descriptor: | Envelope small membrane protein | | Authors: | Zhang, R, Qin, H, Prasad, R, Fu, R, Zhou, H.X, Cross, T. | | Deposit date: | 2023-09-02 | | Release date: | 2023-11-15 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Dimeric Transmembrane Structure of the SARS-CoV-2 E Protein.
Commun Biol, 6, 2023
|
|
8U1S
 
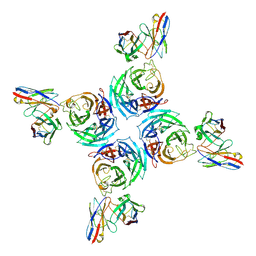 | |
8U1R
 
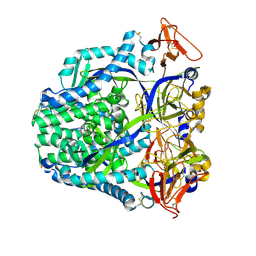 | |
8U1Q
 
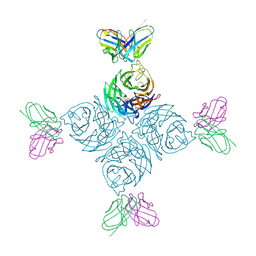 | |
8U1O
 
 | |
8U1N
 
 | |
8U1M
 
 | |
8U1L
 
 | |
8U1K
 
 | | Cryo-EM of Caulobacter crescentus Tad pilus | | Descriptor: | PilA | | Authors: | Sonani, R.R, Sanchez, J.C, Baumgardt, J.K, Wright, E.R, Egelman, E.H. | | Deposit date: | 2023-09-01 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Tad and toxin-coregulated pilus structures reveal unexpected diversity in bacterial type IV pili.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8U1J
 
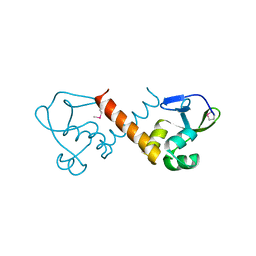 | |
8U1G
 
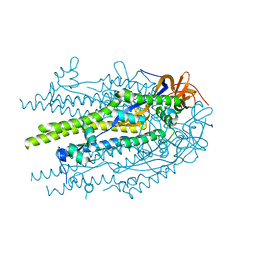 | |
8U1F
 
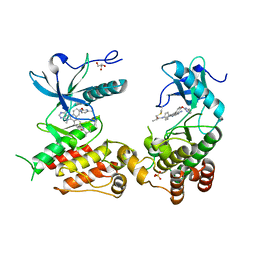 | | FGFR2 Kinase Domain Bound to Irreversible Inhibitor Cmpd 10 | | Descriptor: | Fibroblast growth factor receptor 2, GLYCEROL, N-[4-(4-amino-7-methyl-5-{4-[(4-methylpyrimidin-2-yl)oxy]phenyl}-7H-pyrrolo[2,3-d]pyrimidin-6-yl)phenyl]-2-methylpropanamide, ... | | Authors: | Valverde, R, Foster, L. | | Deposit date: | 2023-08-31 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Discovery of lirafugratinib (RLY-4008), a highly selective irreversible small-molecule inhibitor of FGFR2.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U1E
 
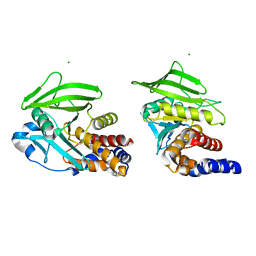 | |
8U1D
 
 | |
8U1C
 
 | |
8U18
 
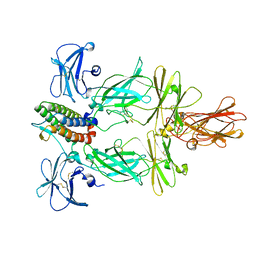 | | Cryo-EM structure of murine Thrombopoietin receptor ectodomain in complex with Tpo | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Thrombopoietin, Thrombopoietin receptor,GCN4 isoform 1, ... | | Authors: | Sarson-Lawrence, K.S, Hardy, J.M, Leis, A, Babon, J.J, Kershaw, N.J. | | Deposit date: | 2023-08-30 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the extracellular domain of murine Thrombopoietin Receptor in complex with Thrombopoietin.
Nat Commun, 15, 2024
|
|
8U17
 
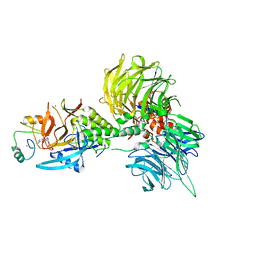 | | The ternary complex structure of DDB1-CRBN-SALL4(ZF1,2)-long bound to Pomalidomide | | Descriptor: | DNA damage-binding protein 1, Protein cereblon, S-Pomalidomide, ... | | Authors: | Clifton, M.C, Ma, X, Ornelas, E. | | Deposit date: | 2023-08-30 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and biophysical comparisons of the pomalidomide- and CC-220-induced interactions of SALL4 with cereblon.
Sci Rep, 13, 2023
|
|
8U16
 
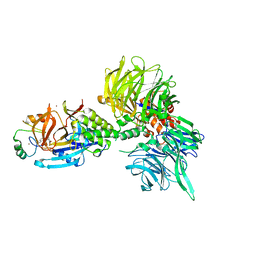 | | The ternary complex structure of DDB1-CRBN-SALL4(ZF1,2)-short bound to Pomalidomide | | Descriptor: | 1,2-ETHANEDIOL, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Clifton, M.C, Ma, X, Ornelas, E. | | Deposit date: | 2023-08-30 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and biophysical comparisons of the pomalidomide- and CC-220-induced interactions of SALL4 with cereblon.
Sci Rep, 13, 2023
|
|
8U15
 
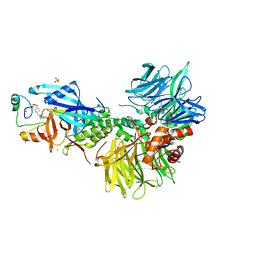 | | The ternary complex structure of DDB1-CRBN-SALL4(ZF1,2)-short bound to CC-220 | | Descriptor: | (3S)-3-[4-({4-[(morpholin-4-yl)methyl]phenyl}methoxy)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DDB1, Protein cereblon, ... | | Authors: | Clifton, M.C, Ma, X, Ornelas, E. | | Deposit date: | 2023-08-30 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural and biophysical comparisons of the pomalidomide- and CC-220-induced interactions of SALL4 with cereblon.
Sci Rep, 13, 2023
|
|
8U14
 
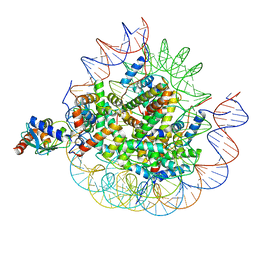 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A lysine 15 in complex with RNF168-UbcH5c (class 2) | | Descriptor: | DNA (146-MER), DNA (147-MER), E3 ubiquitin-protein ligase RNF168, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-08-30 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8U13
 
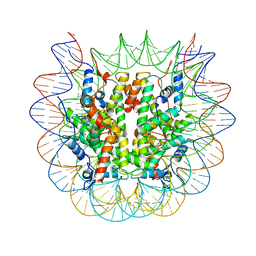 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A lysine 15 in complex with RNF168-UbcH5c (class 1) | | Descriptor: | DNA (146-MER), DNA (147-MER), E3 ubiquitin-protein ligase RNF168, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-08-30 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8U12
 
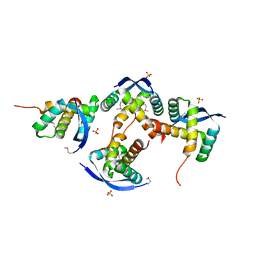 | |
8U11
 
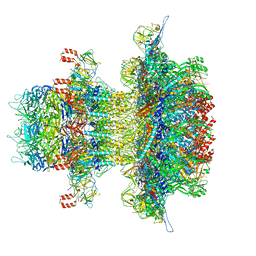 | | In situ cryo-EM structure of bacteriophage P22 gp1:gp5:gp4: gp10: gp9 N-term complex in conformation 2 at 3.1A resolution | | Descriptor: | Major capsid protein, Packaged DNA stabilization protein gp10, Peptidoglycan hydrolase gp4, ... | | Authors: | Iglesias, S, Feng-Hou, C, Cingolani, G. | | Deposit date: | 2023-08-30 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular Architecture of Salmonella Typhimurium Virus P22 Genome Ejection Machinery.
J.Mol.Biol., 435, 2023
|
|
8U10
 
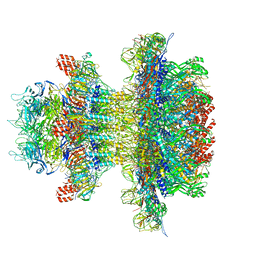 | | In situ cryo-EM structure of bacteriophage P22 gp1:gp4:gp5:gp10:gp9 N-term complex in conformation 1 at 3.2A resolution | | Descriptor: | Major capsid protein, Packaged DNA stabilization protein gp10, Peptidoglycan hydrolase gp4, ... | | Authors: | Iglesias, S, Feng-Hou, C, Cingolani, G. | | Deposit date: | 2023-08-30 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular Architecture of Salmonella Typhimurium Virus P22 Genome Ejection Machinery.
J.Mol.Biol., 435, 2023
|
|
8U0Z
 
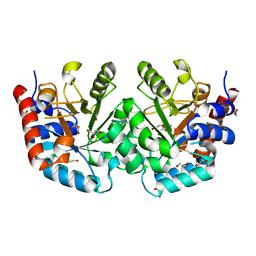 | | CRYSTAL STRUCTURE OF THE OROTIDINE 5'-MONOPHOSPHATE DECARBOXYLASE DOMAIN OF Coffea arabica UMP SYNTHASE | | Descriptor: | 1,2-ETHANEDIOL, ANY 5'-MONOPHOSPHATE NUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hinojosa-Cruz, A, Diaz-Vilchis, A, Gonzalez-Segura, L. | | Deposit date: | 2023-08-29 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Structural and functional properties of uridine 5'-monophosphate synthase from Coffea arabica.
Int.J.Biol.Macromol., 259, 2024
|
|
