3ROS
 
 | |
5CKT
 
 | | Crystal Structure of KorA, a plasmid-encoded, global transcription regulator | | Descriptor: | ACETATE ION, TrfB transcriptional repressor protein | | Authors: | White, S.A, Hyde, E.I, Lovering, A.L. | | Deposit date: | 2015-07-15 | | Release date: | 2016-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Flexibility of KorA, a plasmid-encoded, global transcription regulator, in the presence and the absence of its operator.
Nucleic Acids Res., 44, 2016
|
|
3NRJ
 
 | | Crystal structure of probable yrbi family phosphatase from pseudomonas syringae pv.phaseolica 1448a complexed with magnesium | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Freeman, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-30 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Had Family Hydrolase from Pseudomonas Syringae Pv.Phaseolica 1448A
To be Published
|
|
3WF5
 
 | | Crystal structure of S6K1 kinase domain in complex with a pyrazolopyrimidine derivative 4-[4-(1H-benzimidazol-2-yl)piperidin-1-yl]-1H-pyrazolo[3,4-d]pyrimidine | | Descriptor: | 4-[4-(1H-benzimidazol-2-yl)piperidin-1-yl]-1H-pyrazolo[3,4-d]pyrimidine, Ribosomal protein S6 kinase beta-1, ZINC ION | | Authors: | Niwa, H, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-17 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Crystal structures of the S6K1 kinase domain in complexes with inhibitors
J.Struct.Funct.Genom., 15, 2014
|
|
3NSM
 
 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 from Ostrinia furnacalis | | Descriptor: | N-acetylglucosaminidase | | Authors: | Zhang, H, Liu, T, Liu, F, Yang, Q, Shen, X. | | Deposit date: | 2010-07-02 | | Release date: | 2010-11-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Determinants of an Insect {beta}-N-Acetyl-D-hexosaminidase Specialized as a Chitinolytic Enzyme
J.Biol.Chem., 286, 2011
|
|
3WAN
 
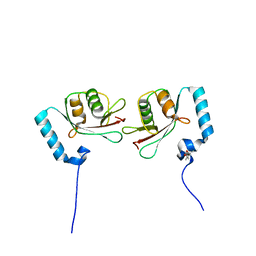 | | Crystal structure of Atg13 LIR-fused human LC3A_2-121 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Autophagy-related protein 13, Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2013-05-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
1BWS
 
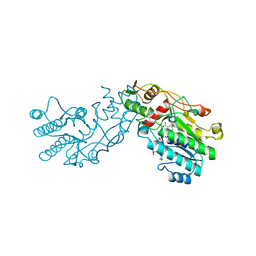 | | CRYSTAL STRUCTURE OF GDP-4-KETO-6-DEOXY-D-MANNOSE EPIMERASE/REDUCTASE FROM ESCHERICHIA COLI A KEY ENZYME IN THE BIOSYNTHESIS OF GDP-L-FUCOSE | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (GDP-4-KETO-6-DEOXY-D-MANNOSE EPIMERASE/REDUCTASE) | | Authors: | Rizzi, M, Tonetti, M, Flora, A.D, Bolognesi, M. | | Deposit date: | 1998-09-25 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | GDP-4-keto-6-deoxy-D-mannose epimerase/reductase from Escherichia coli, a key enzyme in the biosynthesis of GDP-L-fucose, displays the structural characteristics of the RED protein homology superfamily.
Structure, 6, 1998
|
|
5CL9
 
 | |
1V1M
 
 | | CROSSTALK BETWEEN COFACTOR BINDING AND THE PHOSPHORYLATION LOOP CONFORMATION IN THE BCKD MACHINE | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, BENZAMIDINE, ... | | Authors: | Li, J, Wynn, R.M, Machius, M, Chuang, J.L, Karthikeyan, S, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-04-20 | | Release date: | 2004-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cross-Talk between Thiamin Diphosphate Binding and Phosphorylation Loop Conformation in Human Branched-Chain Alpha-Keto Acid Decarboxylase/Dehydrogenase.
J.Biol.Chem., 279, 2004
|
|
4GIC
 
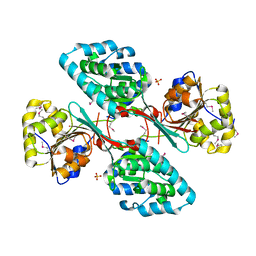 | | Crystal Structure Of a Putative Histidinol dehydrogenase (Target PSI-014034) from Methylococcus capsulatus | | Descriptor: | Histidinol dehydrogenase, SULFATE ION | | Authors: | Kumar, P.R, Ahmed, M, Banu, N, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Gizzi, A, Glen, S, Hammonds, J, Hillerich, B, Love, J.D, Seidel, R, Stead, M, Toro, R, Washington, E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-08-08 | | Release date: | 2012-08-22 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Crystal structure of a putative Histidinol dehydrogenase from Methylococcus capsulatus
to be published
|
|
3RNG
 
 | | Structure of the Toluene/o-Xylene Monooxygenase Hydroxylase T201S/W167E Double Mutant | | Descriptor: | FE (III) ION, HYDROXIDE ION, Toluene o-xylene monooxygenase component | | Authors: | Gucinski, G, Song, W.J, Lippard, S.J, Sazinsky, M.H. | | Deposit date: | 2011-04-22 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Tracking a defined route for O2 migration in a dioxygen-activating diiron enzyme.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8YNR
 
 | |
3WH3
 
 | | human Mincle, ligand free form | | Descriptor: | C-type lectin domain family 4 member E, CALCIUM ION | | Authors: | Furukawa, A, Kamishikiryo, J, Mori, D, Toyonaga, K, Okabe, Y, Toji, A, Kanda, R, Miyake, Y, Ose, T, Yamasaki, S, Maenaka, K. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural analysis for glycolipid recognition by the C-type lectins Mincle and MCL
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3O6A
 
 | | F144Y/F258Y Double Mutant of Exo-beta-1,3-glucanase from Candida albicans at 2 A | | Descriptor: | Glucan 1,3-beta-glucosidase | | Authors: | Nakatani, Y, Cutfield, S.M, Cutfield, J.F. | | Deposit date: | 2010-07-28 | | Release date: | 2010-09-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Carbohydrate binding sites in Candida albicans exo-beta-1,3-glucanase and the role of the Phe-Phe 'clamp' at the active site entrance
Febs J., 277, 2010
|
|
1V3T
 
 | | Crystal structure of leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, leukotriene b4 12-hydroxydehydrogenase/prostaglandin 15-keto reductase | | Authors: | Hori, T, Yokomizo, T, Ago, H, Sugahara, M, Ueno, G, Yamamoto, M, Kumasaka, T, Shimizu, T, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-05 | | Release date: | 2004-07-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of leukotriene B4 12-hydroxydehydrogenase/15-Oxo-prostaglandin 13-reductase catalytic mechanism and a possible Src homology 3 domain binding loop
J.Biol.Chem., 279, 2004
|
|
8ZI2
 
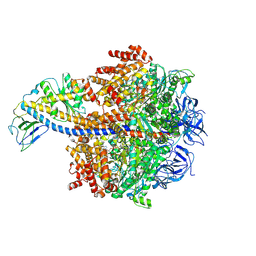 | | Cryo-EM reveals transition states of the Acinetobacter baumannii F1-ATPase rotary subunits gamma and epsilon and novel compound targets - Conformation 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Le, K.C.M, Wong, C.F, Gruber, G. | | Deposit date: | 2024-05-12 | | Release date: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Cryo-EM reveals transition states of the Acinetobacter baumannii F 1 -ATPase rotary subunits gamma and epsilon , unveiling novel compound targets.
Faseb J., 38, 2024
|
|
6T8O
 
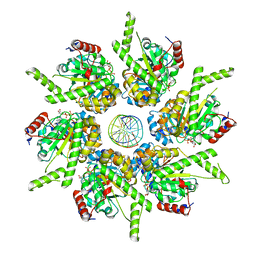 | | Stalled FtsK motor domain bound to dsDNA end | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA translocase FtsK, dsDNA substrate | | Authors: | Jean, N.L, Lowe, J. | | Deposit date: | 2019-10-24 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | FtsK in motion reveals its mechanism for double-stranded DNA translocation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1V5B
 
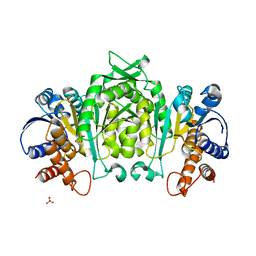 | | The Structure Of The Mutant, S225A and E251L, Of 3-Isopropylmalate Dehydrogenase From Bacillus Coagulans | | Descriptor: | 3-isopropylmalate dehydrogenase, SULFATE ION | | Authors: | Fujita, K, Minami, H, Suzuki, K, Tsunoda, M, Sekiguchi, T, Mizui, R, Tsuzaki, S, Nakamura, S, Takenaka, A. | | Deposit date: | 2003-11-22 | | Release date: | 2005-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of a highly thermo-stabilized mutant of 3-isopropylmalate dehydrogenase from Bacillus coagulans: An evaluation of local packing density in the hydrophobic core
To be Published
|
|
4GFI
 
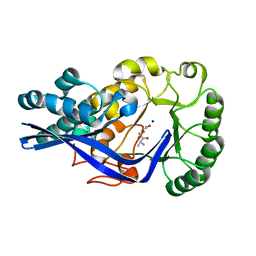 | | Crystal structure of EFI-502318, an enolase family member from Agrobacterium tumefaciens with homology to dipeptide epimerases (bound sodium, L-Ala-L-Glu with ordered loop) | | Descriptor: | ALANINE, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | Vetting, M.W, Bouvier, J.T, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-03 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of EFI-502318, an enolase family member from Agrobacterium tumefaciens with homology to dipeptide epimerases (bound sodium, l-ala-l-glu with ordered loop)
To be Published
|
|
3O7U
 
 | | Crystal structure of Cytosine Deaminase from Escherichia Coli complexed with zinc and phosphono-cytosine | | Descriptor: | (2R)-2-amino-2,5-dihydro-1,5,2-diazaphosphinin-6(1H)-one 2-oxide, (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, Cytosine deaminase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Hall, R.S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2010-07-31 | | Release date: | 2011-06-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.708 Å) | | Cite: | Three-dimensional structure and catalytic mechanism of Cytosine deaminase.
Biochemistry, 50, 2011
|
|
3R1Z
 
 | | Crystal structure of NYSGRC enolase target 200555, a putative dipeptide epimerase from Francisella philomiragia : Complex with L-Ala-L-Glu and L-Ala-D-Glu | | Descriptor: | ALANINE, D-GLUTAMIC ACID, Enzyme of enolase superfamily, ... | | Authors: | Vetting, M.W, Hillerich, B, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-03-11 | | Release date: | 2011-04-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1V6U
 
 | | Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 2(2)-alpha-L-arabinofuranosyl-xylobiose | | Descriptor: | alpha-D-xylopyranose, alpha-L-arabinofuranose-(1-3)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, beta-D-xylopyranose, ... | | Authors: | Fujimoto, Z, Kaneko, S, Kuno, A, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2003-12-04 | | Release date: | 2004-04-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of decorated xylooligosaccharides bound to a family 10 xylanase from Streptomyces olivaceoviridis E-86
J.Biol.Chem., 279, 2004
|
|
1UXV
 
 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-03-01 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
6TA6
 
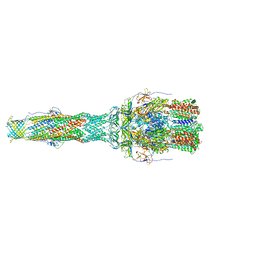 | | MexAB assembly of the Pseudomonas MexAB-OprM efflux pump reconstituted in nanodiscs | | Descriptor: | Efflux pump membrane transporter, MexA family multidrug efflux RND transporter periplasmic adaptor subunit, Outer membrane protein OprM | | Authors: | Glavier, M, Schoehn, G, Taveau, J.C, Phan, G, Daury, L, Lambert, O, Broutin, I. | | Deposit date: | 2019-10-29 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Antibiotic export by MexB multidrug efflux transporter is allosterically controlled by a MexA-OprM chaperone-like complex.
Nat Commun, 11, 2020
|
|
8YP0
 
 | |
