1DOR
 
 | | DIHYDROOROTATE DEHYDROGENASE A FROM LACTOCOCCUS LACTIS | | Descriptor: | DIHYDROOROTATE DEHYDROGENASE A, FLAVIN MONONUCLEOTIDE | | Authors: | Rowland, P, Larsen, S. | | Deposit date: | 1997-01-14 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the flavin containing enzyme dihydroorotate dehydrogenase A from Lactococcus lactis.
Structure, 5, 1997
|
|
6I26
 
 | | Rea1 Wild type AMPPNP state | | Descriptor: | Midasin,Midasin,Midasin,Midasin | | Authors: | Sosnowski, P, Urnavicius, L, Boland, A, Fagiewicz, R, Busselez, J, Papai, G, Schmidt, H. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | The CryoEM structure of the Saccharomyces cerevisiae ribosome maturation factor Rea1.
Elife, 7, 2018
|
|
5A5D
 
 | | A complex of the synthetic siderophore analogue Fe(III)-5-LICAM with the CeuE periplasmic protein from Campylobacter jejuni | | Descriptor: | ENTEROCHELIN UPTAKE PERIPLASMIC BINDING PROTEIN, FE (III) ION, N,N'-pentane-1,5-diylbis(2,3-dihydroxybenzamide) | | Authors: | Blagova, E, Hughes, A, Moroz, O.V, Raines, D.J, Wilde, E.J, Turkenburg, J.P, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2015-06-17 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
3KIA
 
 | | Crystal structure of mannosyl-3-phosphoglycerate synthase from Rubrobacter xylanophilus | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Macedo-Ribeiro, S, Pereira, P.J.B, Empadinhas, N, da Costa, M.S. | | Deposit date: | 2009-11-01 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Functional and structural characterization of a novel mannosyl-3-phosphoglycerate synthase from Rubrobacter xylanophilus reveals its dual substrate specificity
Mol.Microbiol., 79, 2011
|
|
1ISZ
 
 | | Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with galactose | | Descriptor: | beta-D-galactopyranose, endo-1,4-beta-D-xylanase | | Authors: | Fujimoto, Z, Kuno, A, Kaneko, S, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2001-12-27 | | Release date: | 2002-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase: sugar binding structure of the family 13 carbohydrate binding module.
J.Mol.Biol., 316, 2002
|
|
1YX9
 
 | |
1IWU
 
 | | Crystal Structure Analysis of Human lysozyme at 127K. | | Descriptor: | CHLORIDE ION, LYSOZYME C | | Authors: | Joti, Y, Nakasako, M, Kidera, A, Go, N. | | Deposit date: | 2002-06-03 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Nonlinear temperature dependence of the crystal structure of lysozyme: correlation between coordinate shifts and thermal factors.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1IUV
 
 | | P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-4-HYDROXYBENZOATE AT PH 5.0 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID | | Authors: | Gatti, D.L, Entsch, B, Ballou, D.P, Ludwig, M.L. | | Deposit date: | 1995-11-22 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | pH-dependent structural changes in the active site of p-hydroxybenzoate hydroxylase point to the importance of proton and water movements during catalysis.
Biochemistry, 35, 1996
|
|
1YYN
 
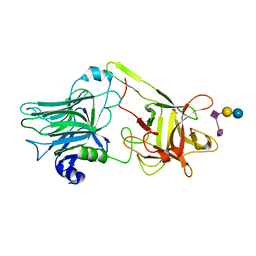 | | A common binding site for disialyllactose and a tri-peptide in the C-fragment of tetanus neurotoxin | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-3)-alpha-D-galactopyranose-(1-4)-beta-D-glucopyranose, Tetanus toxin | | Authors: | Seetharaman, J, Eswaramoorthy, S, Kumaran, D, Swaminathan, S. | | Deposit date: | 2005-02-25 | | Release date: | 2005-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Common binding site for disialyllactose and tri-peptide in C-fragment of tetanus neurotoxin
Proteins, 61, 2005
|
|
1IVM
 
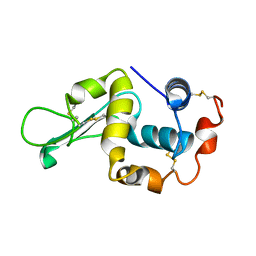 | |
1Y6W
 
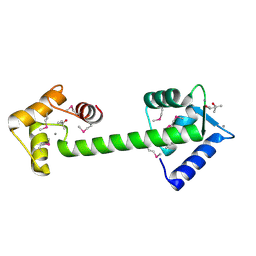 | | Trapped intermediate of calmodulin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Calmodulin, ... | | Authors: | Grabarek, Z. | | Deposit date: | 2004-12-07 | | Release date: | 2005-03-01 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a Trapped Intermediate of Calmodulin: Calcium Regulation of EF-hand Proteins from a New Perspective.
J.Mol.Biol., 346, 2005
|
|
5CRV
 
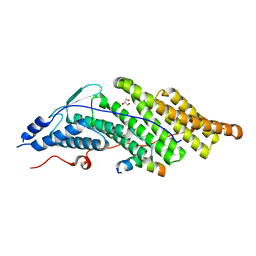 | | Crystal structure of the Bro domain of HD-PTP in a complex with the core region of STAM2 | | Descriptor: | GLYCEROL, Signal transducing adapter molecule 2, Tyrosine-protein phosphatase non-receptor type 23 | | Authors: | Lee, J, Ku, B, Kim, S.J. | | Deposit date: | 2015-07-23 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural Study of the HD-PTP Bro1 Domain in a Complex with the Core Region of STAM2, a Subunit of ESCRT-0
Plos One, 11, 2016
|
|
1KX5
 
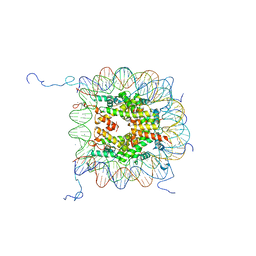 | | X-Ray Structure of the Nucleosome Core Particle, NCP147, at 1.9 A Resolution | | Descriptor: | CHLORIDE ION, DNA (5'(ATCAATATCCACCTGCAGATACTACCAAAAGTGTATTTGGAAACTGCTCCATCAAAAGGCATGTTCAGCTGGAATCCAGCTGAACATGCCTTTTGATGGAGCAGTTTCCAAATACACTTTTGGTAGTATCTGCAGGTGGATATTGAT)3'), DNA (5'(ATCAATATCCACCTGCAGATACTACCAAAAGTGTATTTGGAAACTGCTCCATCAAAAGGCATGTTCAGCTGGATTCCAGCTGAACATGCCTTTTGATGGAGCAGTTTCCAAATACACTTTTGGTAGTATCTGCAGGTGGATATTGAT)3'), ... | | Authors: | Davey, C.A, Sargent, D.F, Luger, K, Maeder, A.W, Richmond, T.J. | | Deposit date: | 2002-01-31 | | Release date: | 2002-12-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Solvent Mediated Interactions in the Structure of the Nucleosome Core Particle at 1.9 A Resolution
J.Mol.Biol., 319, 2002
|
|
1KXQ
 
 | | Camelid VHH Domain in Complex with Porcine Pancreatic alpha-Amylase | | Descriptor: | CALCIUM ION, CHLORIDE ION, alpha-amylase, ... | | Authors: | Desmyter, A, Spinelli, S, Payan, F, Lauwereys, M, Wyns, L, Muyldermans, S, Cambillau, C. | | Deposit date: | 2002-02-01 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Three camelid VHH domains in complex with porcine pancreatic alpha-amylase. Inhibition and versatility of binding topology.
J.Biol.Chem., 277, 2002
|
|
6CVY
 
 | | Crystal structure of HCV NS3/4A WT protease in complex with AJ-21 (MK-5172 linear analogue) | | Descriptor: | 3-methyl-N-[(pentyloxy)carbonyl]-L-valyl-(4R)-4-[(3-chloro-7-methoxyquinoxalin-2-yl)oxy]-N-[(1R,2S)-2-ethenyl-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclopropyl]-L-prolinamide, GLYCEROL, NS3 protease, ... | | Authors: | Matthew, A.N, Schiffer, C.A. | | Deposit date: | 2018-03-29 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Quinoxaline-Based Linear HCV NS3/4A Protease Inhibitors Exhibit Potent Activity against Drug Resistant Variants.
ACS Med Chem Lett, 9, 2018
|
|
1KVS
 
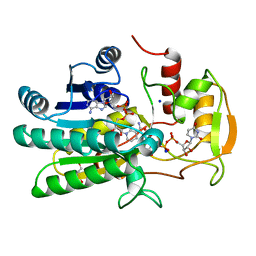 | | UDP-GALACTOSE 4-EPIMERASE COMPLEXED WITH UDP-PHENOL | | Descriptor: | DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Thoden, J.B, Gulick, A.M, Holden, H.M. | | Deposit date: | 1997-03-07 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular structures of the S124A, S124T, and S124V site-directed mutants of UDP-galactose 4-epimerase from Escherichia coli.
Biochemistry, 36, 1997
|
|
3V03
 
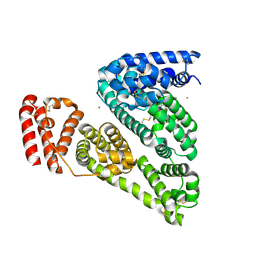 | | Crystal structure of Bovine Serum Albumin | | Descriptor: | ACETATE ION, CALCIUM ION, Serum albumin | | Authors: | Majorek, K.A, Porebski, P.J, Chruszcz, M, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-07 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and immunologic characterization of bovine, horse, and rabbit serum albumins.
Mol.Immunol., 52, 2012
|
|
1KVR
 
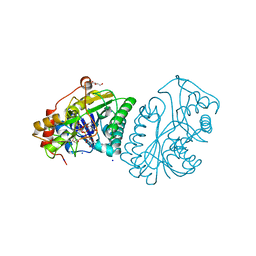 | | UDP-GALACTOSE 4-EPIMERASE COMPLEXED WITH UDP-PHENOL | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Thoden, J.B, Gulick, A.M, Holden, H.M. | | Deposit date: | 1997-03-07 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular structures of the S124A, S124T, and S124V site-directed mutants of UDP-galactose 4-epimerase from Escherichia coli.
Biochemistry, 36, 1997
|
|
1KVU
 
 | | UDP-GALACTOSE 4-EPIMERASE COMPLEXED WITH UDP-PHENOL | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Thoden, J.B, Gulick, A.M, Holden, H.M. | | Deposit date: | 1997-03-07 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic roles of tyrosine 149 and serine 124 in UDP-galactose 4-epimerase from Escherichia coli.
Biochemistry, 36, 1997
|
|
6I3K
 
 | | Bilirubin oxidase from Myrothecium verrucaria, mutant W396A in complex with ferricyanide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bilirubin oxidase, COPPER (II) ION, ... | | Authors: | Koval, T, Svecova, L, Skalova, T, Kolenko, P, Duskova, J, Ostergaard, L.H, Dohnalek, J. | | Deposit date: | 2018-11-06 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Trp-His covalent adduct in bilirubin oxidase is crucial for effective bilirubin binding but has a minor role in electron transfer.
Sci Rep, 9, 2019
|
|
3HEI
 
 | | Ligand Recognition by A-Class Eph Receptors: Crystal Structures of the EphA2 Ligand-Binding Domain and the EphA2/ephrin-A1 Complex | | Descriptor: | Ephrin type-A receptor 2, Ephrin-A1 | | Authors: | Himanen, J.P, Goldgur, Y, Miao, H, Myshkin, E, Guo, H, Buck, M, Nguyen, M, Rajashankar, K.R, Wang, B, Nikolov, D.B. | | Deposit date: | 2009-05-08 | | Release date: | 2009-06-30 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand recognition by A-class Eph receptors: crystal structures of the EphA2 ligand-binding domain and the EphA2/ephrin-A1 complex.
Embo Rep., 10, 2009
|
|
4MW1
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(3-chloro-5-methoxybenzyl)amino]propyl}-3-thiophen-3-ylurea (Chem 1444) | | Descriptor: | 1-{3-[(3-chloro-5-methoxybenzyl)amino]propyl}-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
1DOC
 
 | | THE MOBIL FLAVIN OF 4-OH BENZOATE HYDROXYLASE: MOTION OF A PROSTHETIC GROUP REGULATES CATALYSIS | | Descriptor: | BROMIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, ... | | Authors: | Gatti, D.L, Palfey, B.A, Lah, M.S, Entsch, B, Massey, V, Ballou, D.P, Ludwig, M.L. | | Deposit date: | 1994-09-06 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The mobile flavin of 4-OH benzoate hydroxylase.
Science, 266, 1994
|
|
6CUH
 
 | | Crystal structure of the unliganded BC8B TCR | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shahine, A.E, Rossjohn, J. | | Deposit date: | 2018-03-26 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A T-cell receptor escape channel allows broad T-cell response to CD1b and membrane phospholipids.
Nat Commun, 10, 2019
|
|
1Q41
 
 | | GSK-3 Beta complexed with Indirubin-3'-monoxime | | Descriptor: | (Z)-1H,1'H-[2,3']BIINDOLYLIDENE-3,2'-DIONE-3-OXIME, GLYCOGEN SYNTHASE KINASE-3 BETA | | Authors: | Bertrand, J.A, Thieffine, S, Vulpetti, A, Cristiani, C, Valsasina, B, Knapp, S, Kalisz, H.M, Flocco, M. | | Deposit date: | 2003-08-01 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of the Gsk-3Beta Active Site Using Selective and Non-selective ATP-Mimetic Inhibitors
J.Mol.Biol., 333, 2003
|
|
