5IIJ
 
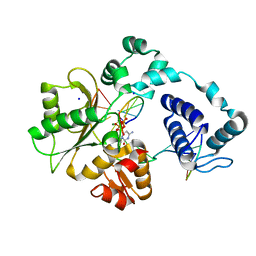 | | Crystal structure of the pre-catalytic ternary complex of DNA polymerase lambda with a templating 8-oxo-dG and an incoming dCTP | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA (5'-D(*CP*AP*GP*TP*AP*(DOC))-3'), DNA (5'-D(*CP*GP*GP*CP*(8OG)P*GP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Burak, M.J, Guja, K.E, Garcia-Diaz, M. | | Deposit date: | 2016-03-01 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.724 Å) | | Cite: | A fidelity mechanism in DNA polymerase lambda promotes error-free bypass of 8-oxo-dG.
Embo J., 35, 2016
|
|
5WA8
 
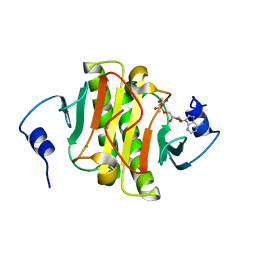 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant nucleoside L-Ala phosphoramidate substrate complex | | Descriptor: | CHLORIDE ION, Histidine triad nucleotide-binding protein 1, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-~{N}-[(2~{S})-1-methoxy-1-oxidanylidene-propan-2-yl]phosphonamidic acid | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2017-06-26 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated ProTides.
Mol. Pharm., 14, 2017
|
|
5WMQ
 
 | |
6I1D
 
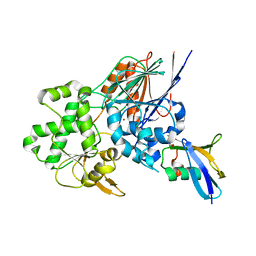 | | Structure of the Ysh1-Mpe1 nuclease complex from S.cerevisiae | | Descriptor: | Endoribonuclease YSH1, GLYCEROL, Protein MPE1, ... | | Authors: | Hill, C.H, Boreikaite, V, Kumar, A, Casanal, A, Kubik, P, Degliesposti, G, Maslen, S, Mariani, A, von Loeffelholz, O, Girbig, M, Skehel, M, Passmore, L.A. | | Deposit date: | 2018-10-28 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Activation of the Endonuclease that Defines mRNA 3' Ends Requires Incorporation into an 8-Subunit Core Cleavage and Polyadenylation Factor Complex.
Mol.Cell, 73, 2019
|
|
5IKY
 
 | |
5WMW
 
 | | Structural Insights into Substrate and Inhibitor Binding Sites in Human Indoleamine 2,3-Dioxygenase 1 | | Descriptor: | CYANIDE ION, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lewis-Ballester, A, Yeh, S.R, Pham, K.N, Batabyal, D, Karkashon, S, Bonanno, J.B, Poulos, T.L. | | Deposit date: | 2017-07-31 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structural insights into substrate and inhibitor binding sites in human indoleamine 2,3-dioxygenase 1.
Nat Commun, 8, 2017
|
|
5WCR
 
 | | Phosphotriesterase variant R0deltaL7 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-R0, ... | | Authors: | Miton, C.M, Campbell, E.C, Jackson, C.J, Tokuriki, N. | | Deposit date: | 2017-07-01 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Phosphotriesterase variant R0deltaL7
To Be Published
|
|
5ILE
 
 | | H64A sperm whale myoglobin with a Fe-tolyl moiety | | Descriptor: | Myoglobin, SULFATE ION, [3,3'-(7,12-diethenyl-3,8,13,17-tetramethylporphyrin-2,18-diyl-kappa~4~N~21~,N~22~,N~23~,N~24~)di(propanoato)(2-)](3-methylphenyl)iron | | Authors: | Wang, B, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2016-03-03 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Organometallic myoglobins: Formation of Fe-carbon bonds and distal pocket effects on aryl ligand conformations.
J. Inorg. Biochem., 164, 2016
|
|
6I1I
 
 | | Crystal structure of TP domain from Escherichia coli penicillin-binding protein 3 in complex with penicillin | | Descriptor: | Peptidoglycan D,D-transpeptidase FtsI,Peptidoglycan D,D-transpeptidase FtsI, Piperacillin (Open Form) | | Authors: | Bellini, D, Koekemoer, L, Newman, H, Dowson, C.G. | | Deposit date: | 2018-10-28 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Novel and Improved Crystal Structures of H. influenzae, E. coli and P. aeruginosa Penicillin-Binding Protein 3 (PBP3) and N. gonorrhoeae PBP2: Toward a Better Understanding of beta-Lactam Target-Mediated Resistance.
J.Mol.Biol., 431, 2019
|
|
5WO2
 
 | | Chaperone Spy bound to Casein Fragment (Casein un-modeled) | | Descriptor: | CHLORIDE ION, IMIDAZOLE, Periplasmic chaperone Spy, ... | | Authors: | Horowitz, S, Koldewey, P, Martin, R, Bardwell, J.C.A. | | Deposit date: | 2017-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | Visualizing chaperone-assisted protein folding.
Nat. Struct. Mol. Biol., 23, 2016
|
|
6I5X
 
 | | Crystal structure of Aspergillus fumigatus phosphomannomutase | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Zhang, Y, Raimi, O.G, Ferenbach, A.T, van Aalten, D.M.F. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Aspergillus fumigatus phosphomannomutase
To Be Published
|
|
5WEW
 
 | | Crystal structure of Klebsiella pneumoniae fosfomycin resistance protein (FosAKP) with inhibitor (ANY1) bound | | Descriptor: | 6,6'-(4-nitro-1H-pyrazole-3,5-diyl)bis(3-bromopyrazolo[1,5-a]pyrimidin-2(1H)-one), Fosfomycin resistance protein, MANGANESE (II) ION | | Authors: | Klontz, E.H, Sundberg, E.J. | | Deposit date: | 2017-07-10 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.178 Å) | | Cite: | Small-Molecule Inhibitor of FosA Expands Fosfomycin Activity to Multidrug-Resistant Gram-Negative Pathogens.
Antimicrob. Agents Chemother., 63, 2019
|
|
6I22
 
 | | Flavin Analogue Sheds Light on Light-Oxygen-Voltage Domain Mechanism | | Descriptor: | Aureochrome1-like protein, FLAVIN MONONUCLEOTIDE | | Authors: | Rizkallah, P.J, Kalvaitis, M.E, Allemann, R.K, Mart, R.J, Johnson, L.A. | | Deposit date: | 2018-10-31 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | A Noncanonical Chromophore Reveals Structural Rearrangements of the Light-Oxygen-Voltage Domain upon Photoactivation.
Biochemistry, 58, 2019
|
|
5WPB
 
 | | Crystal structure of fragment 3-(3-(pyridin-2-ylmethoxy)quinoxalin-2-yl)propanoic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-{3-[(pyridin-2-yl)methoxy]quinoxalin-2-yl}propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Tempel, W, Ferreira de Freitas, R, Franzoni, I, Ravichandran, M, Lautens, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-04 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Small Molecule Antagonists of the Interaction between the Histone Deacetylase 6 Zinc-Finger Domain and Ubiquitin.
J. Med. Chem., 60, 2017
|
|
5INI
 
 | | Structural basis for acyl-CoA carboxylase-mediated assembly of unusual polyketide synthase extender units incorporated into the stambomycin antibiotics | | Descriptor: | HEXANOYL-COENZYME A, Putative carboxyl transferase | | Authors: | Valentic, T.R, Ray, L, Miyazawa, T, Withall, D.M, Song, L, Milligan, J.C, Osada, H, Tsai, S.C, Challis, G.L. | | Deposit date: | 2016-03-07 | | Release date: | 2016-12-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A crotonyl-CoA reductase-carboxylase independent pathway for assembly of unusual alkylmalonyl-CoA polyketide synthase extender units.
Nat Commun, 7, 2016
|
|
6I2P
 
 | | Crystal structure of the Mycobacterium tuberculosis PknB kinase domain (L33E mutant) in complex with its substrate GarA | | Descriptor: | Glycogen accumulation regulator GarA, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Andre-Leroux, G, Hindie, V, Barilone, N, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2018-11-01 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural insights into the functional versatility of an FHA domain protein in mycobacterial signaling.
Sci.Signal., 12, 2019
|
|
5WPK
 
 | |
5WGR
 
 | | Crystal Structure of Wild-type MalA', premalbrancheamide complex | | Descriptor: | (5aS,12aS,13aS)-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Fraley, A.E, Smith, J.L. | | Deposit date: | 2017-07-14 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.362 Å) | | Cite: | Function and Structure of MalA/MalA', Iterative Halogenases for Late-Stage C-H Functionalization of Indole Alkaloids.
J. Am. Chem. Soc., 139, 2017
|
|
5I5J
 
 | |
5WGZ
 
 | | Crystal Structure of Wild-type MalA', isomalbrancheamide B complex | | Descriptor: | (5aS,12aS,13aS)-8-chloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Fraley, A.E, Smith, J.L. | | Deposit date: | 2017-07-14 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.041 Å) | | Cite: | Function and Structure of MalA/MalA', Iterative Halogenases for Late-Stage C-H Functionalization of Indole Alkaloids.
J. Am. Chem. Soc., 139, 2017
|
|
6I5Y
 
 | | Crystal structure of E. coli tyrRS in complex with 5'-O-(N-L-tyrosyl)sulfamoyl-adenosine | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[N-(L-TYROSYL)SULFAMOYL]ADENOSINE, Tyrosine--tRNA ligase | | Authors: | De Graef, S, Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2018-11-15 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparative analysis of pyrimidine substituted aminoacyl-sulfamoyl nucleosides as potential inhibitors targeting class I aminoacyl-tRNA synthetases.
Eur.J.Med.Chem., 173, 2019
|
|
6I6I
 
 | | Circular permutant of ribosomal protein S6, adding 6aa to C terminal of P68-69, L75A mutant | | Descriptor: | 30S ribosomal protein S6,30S ribosomal protein S6, SULFATE ION | | Authors: | Wang, H, Logan, D.T, Oliveberg, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Exposing the distinctive modular behavior of beta-strands and alpha-helices in folded proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6I6M
 
 | | Papaver somniferum O-methyltransferase 1 | | Descriptor: | (13aS)-3,10-dimethoxy-5,8,13,13a-tetrahydro-6H-isoquino[3,2-a]isoquinoline-2,9-diol, O-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Cabry, M.P, Offen, W.A, Winzer, T, Li, Y, Graham, I.A, Davies, G.J, Saleh, P. | | Deposit date: | 2018-11-15 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of Papaver somniferum O-Methyltransferase 1 Reveals Initiation of Noscapine Biosynthesis with Implications for Plant Natural Product Methylation
Acs Catalysis, 2019
|
|
5WJ4
 
 | |
6I8V
 
 | | KtrC with ATP bound | | Descriptor: | 1,4-BUTANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Rocha, R, Morais-Cabral, J.H. | | Deposit date: | 2018-11-21 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | Characterization of the molecular properties of KtrC, a second RCK domain that regulates a Ktr channel in Bacillus subtilis.
J.Struct.Biol., 205, 2019
|
|
