5QCM
 
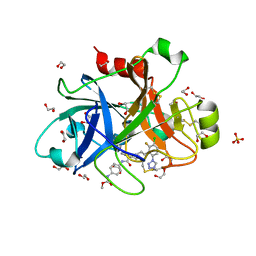 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl ~{N}-[4-[[(1~{S})-2-[(~{E})-3-[3-chloranyl-2-fluoranyl-6-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-3,4-dihydro-1~{H}-isoquinolin-1-yl]carbonylamino]phenyl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Parenteral Small Molecule Coagulation Factor XIa Inhibitor Clinical Candidate (BMS-962212).
J. Med. Chem., 60, 2017
|
|
5AWR
 
 | |
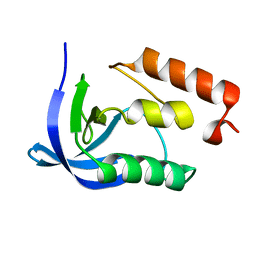
1EY4
 
 | |
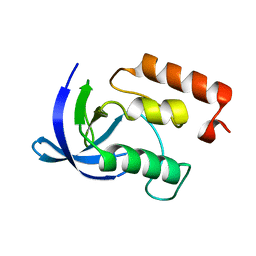
1EYA
 
 | |
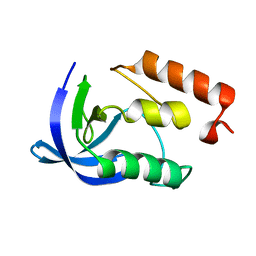
1EY5
 
 | |
1EY9
 
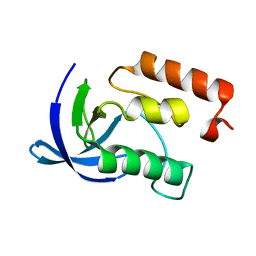 | |
5ELO
 
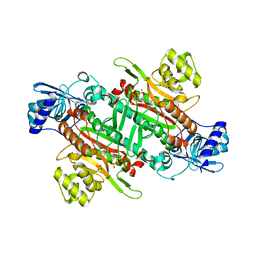 | |
5QCL
 
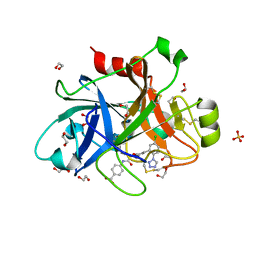 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR 4-[[(1~{S})-2-[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-3,4-dihydro-1~{H}-isoquinolin-1-yl]carbonylamino]benzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-[[(1~{S})-2-[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-3,4-dihydro-1~{H}-isoquinolin-1-yl]carbonylamino]benzoic acid, Coagulation factor XI, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Discovery of a Parenteral Small Molecule Coagulation Factor XIa Inhibitor Clinical Candidate (BMS-962212).
J. Med. Chem., 60, 2017
|
|
6ZNC
 
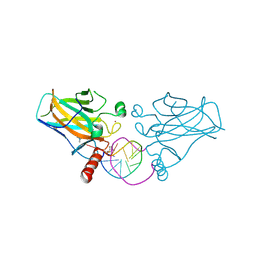 | | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ). Human wild-type p53DBD bound to DNA and MQ: wt-DNA-MQ (I) | | Descriptor: | (2~{R})-2-methyl-1-azabicyclo[2.2.2]octan-3-one, (2~{S})-2-methyl-1-azabicyclo[2.2.2]octan-3-one, Cellular tumor antigen p53, ... | | Authors: | Rozenberg, H, Degtjarik, O, Diskin-Posner, Y, Shakked, Z. | | Deposit date: | 2020-07-06 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ).
Nat Commun, 12, 2021
|
|
5ZG3
 
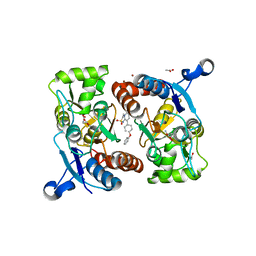 | | Crystal structure of the GluA2o LBD in complex with glutamate and TAK-137 | | Descriptor: | 9-(4-phenoxyphenyl)-3,4-dihydro-2H-2lambda~6~-pyrido[2,1-c][1,2,4]thiadiazine-2,2-dione, ACETATE ION, GLUTAMIC ACID, ... | | Authors: | Sogabe, S, Igaki, S, Hirokawa, A, Zama, Y, Lane, W, Snell, G. | | Deposit date: | 2018-03-07 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | TAK-137, an AMPA-R potentiator with little agonistic effect, has a wide therapeutic window.
Neuropsychopharmacology, 44, 2019
|
|
6PV8
 
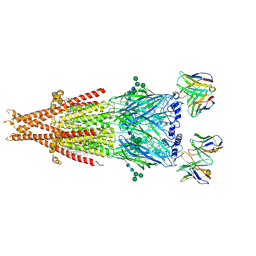 | | Human alpha3beta4 nicotinic acetylcholine receptor in complex with AT-1001 | | Descriptor: | (3-endo)-N-(2-bromophenyl)-9-methyl-9-azabicyclo[3.3.1]nonan-3-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gharpure, A, Teng, J, Zhuang, Y, Noviello, C.M, Walsh, R.M, Cabuco, R, Howard, R.J, Zaveri, N.T, Lindahl, E, Hibbs, R.E. | | Deposit date: | 2019-07-19 | | Release date: | 2019-09-11 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Agonist Selectivity and Ion Permeation in the alpha 3 beta 4 Ganglionic Nicotinic Receptor.
Neuron, 104, 2019
|
|
3EFX
 
 | | Novel binding site identified in a hybrid between cholera toxin and heat-labile enterotoxin, 1.9A crystal structure reveals the details | | Descriptor: | Cholera enterotoxin subunit B, Heat-labile enterotoxin B chain, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]beta-D-glucopyranose | | Authors: | Holmner, A, Lebens, M, Teneberg, S, Angstrom, J, Okvist, M, Krengel, U. | | Deposit date: | 2008-09-10 | | Release date: | 2008-09-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Novel binding site identified in a hybrid between cholera toxin and heat-labile enterotoxin: 1.9 A crystal structure reveals the details
Structure, 12, 2004
|
|
8PQM
 
 | |
1A78
 
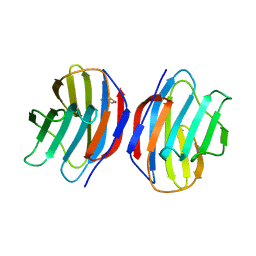 | | COMPLEX OF TOAD OVARY GALECTIN WITH THIO-DIGALACTOSE | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GALECTIN-1, beta-D-galactopyranose-(1-1)-1-thio-beta-D-galactopyranose | | Authors: | Amzel, L.M, Bianchet, M.A, Ahmed, H, Vasta, G.R. | | Deposit date: | 1998-03-20 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Soluble beta-galactosyl-binding lectin (galectin) from toad ovary: crystallographic studies of two protein-sugar complexes
Proteins, 40, 2000
|
|
6PV7
 
 | | Human alpha3beta4 nicotinic acetylcholine receptor in complex with nicotine | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gharpure, A, Teng, J, Zhuang, Y, Noviello, C.M, Walsh, R.M, Cabuco, R, Howard, R.J, Zaveri, N.T, Lindahl, E, Hibbs, R.E. | | Deposit date: | 2019-07-19 | | Release date: | 2019-09-11 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Agonist Selectivity and Ion Permeation in the alpha 3 beta 4 Ganglionic Nicotinic Receptor.
Neuron, 104, 2019
|
|
2WN9
 
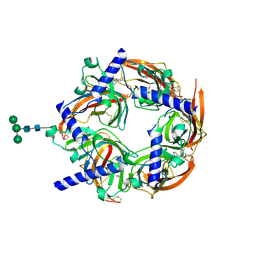 | | Crystal structure of Aplysia ACHBP in complex with 4-0H-DMXBA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(E)-5,6-DIHYDRO-2,3'-BIPYRIDIN-3(4H)-YLIDENEMETHYL]-3-METHOXYPHENOL, SOLUBLE ACETYLCHOLINE RECEPTOR, ... | | Authors: | Sulzenbacher, G, Hibbs, R, Shi, J, Talley, T, Conrod, S, Kem, W, Taylor, P, Marchot, P, Bourne, Y. | | Deposit date: | 2009-07-07 | | Release date: | 2009-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural determinants for interaction of partial agonists with acetylcholine binding protein and neuronal alpha7 nicotinic acetylcholine receptor.
Embo J., 28, 2009
|
|
6S98
 
 | |
3FTC
 
 | |
1GT5
 
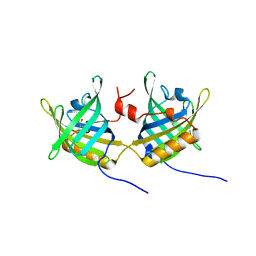 | | Complexe of Bovine Odorant Binding Protein with benzophenone | | Descriptor: | DIPHENYLMETHANONE, ODORANT-BINDING PROTEIN | | Authors: | Vincent, F, Ramoni, R, Spinelli, S, Grolli, S, Conti, V, Cambillau, C, Tegoni, M. | | Deposit date: | 2002-01-10 | | Release date: | 2003-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structures of Bovine Odorant-Binding Protein in Complex with Odorant Molecules.
Eur.J.Biochem., 271, 2004
|
|
1CYJ
 
 | | CYTOCHROME C6 | | Descriptor: | CADMIUM ION, CYTOCHROME C6, HEME C | | Authors: | Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 1995-05-09 | | Release date: | 1996-01-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of chloroplast cytochrome c6 at 1.9 A resolution: evidence for functional oligomerization.
J.Mol.Biol., 250, 1995
|
|
4DBG
 
 | | Crystal structure of HOIL-1L-UBL complexed with a HOIP-UBA derivative | | Descriptor: | RING finger protein 31, RanBP-type and C3HC4-type zinc finger-containing protein 1 | | Authors: | Yagi, H, Hiromoto, T, Mizushima, T, Kurimoto, E, Kato, K. | | Deposit date: | 2012-01-15 | | Release date: | 2012-04-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | A non-canonical UBA-UBL interaction forms the linear-ubiquitin-chain assembly complex
Embo Rep., 13, 2012
|
|
8SZK
 
 | |
1YDR
 
 | | STRUCTURE OF CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT IN COMPLEX WITH H7 PROTEIN KINASE INHIBITOR 1-(5-ISOQUINOLINESULFONYL)-2-METHYLPIPERAZINE | | Descriptor: | 1-(5-ISOQUINOLINESULFONYL)-2-METHYLPIPERAZINE, C-AMP-DEPENDENT PROTEIN KINASE, PROTEIN KINASE INHIBITOR PEPTIDE | | Authors: | Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | Deposit date: | 1996-07-24 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8, and H89. Structural implications for selectivity.
J.Biol.Chem., 271, 1996
|
|
1YDT
 
 | | STRUCTURE OF CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT IN COMPLEX WITH H89 PROTEIN KINASE INHIBITOR N-[2-(4-BROMOCINNAMYLAMINO)ETHYL]-5-ISOQUINOLINE | | Descriptor: | C-AMP-DEPENDENT PROTEIN KINASE, N-[2-(4-BROMOCINNAMYLAMINO)ETHYL]-5-ISOQUINOLINE SULFONAMIDE, PROTEIN KINASE INHIBITOR PEPTIDE | | Authors: | Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | Deposit date: | 1996-07-24 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8, and H89. Structural implications for selectivity.
J.Biol.Chem., 271, 1996
|
|
1YDS
 
 | | Structure of CAMP-dependent protein kinase, alpha-catalytic subunit in complex with H8 protein kinase inhibitor [N-(2-methylamino)ethyl]-5-isoquinolinesulfonamide | | Descriptor: | C-AMP-DEPENDENT PROTEIN KINASE, N-[2-(METHYLAMINO)ETHYL]-5-ISOQUINOLINESULFONAMIDE, PROTEIN KINASE INHIBITOR PEPTIDE | | Authors: | Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | Deposit date: | 1996-07-24 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8, and H89. Structural implications for selectivity.
J.Biol.Chem., 271, 1996
|
|
