1W7W
 
 | |
1W7X
 
 | | Factor7 - 413 complex | | Descriptor: | (S)-[(R)-2-(4-BENZYLOXY-3-METHOXY-PHENYL)-2-(4-CARBAMIMIDOYL-PHENYLAMINO)-ACETYLAMINO]-PHENYL-ACETIC ACID, BLOOD COAGULATION FACTOR VIIA, CALCIUM ION, ... | | Authors: | Ackermann, J, Alig, L, Banner, D.W, Boehm, H.-J, Groebke-Zbinden, K, Hilpert, K, Lave, T, Kuehne, H, Obst-Sander, U, Riederer, M.A, Stahl, M, Tschopp, T.B, Weber, L, Wessel, H.P. | | Deposit date: | 2004-09-14 | | Release date: | 2005-10-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Selective and Orally Bioavailable Phenylglycine Tissue Factor/Factor Viia Inhibitors.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1W7Z
 
 | | Crystal structure of the free (uncomplexed) Ecballium elaterium trypsin inhibitor (EETI-II) | | Descriptor: | FORMIC ACID, SODIUM ION, TRYPSIN INHIBITOR II | | Authors: | Kraetzner, R, Debreczeni, J.E, Pape, T, Kolmar, H, Schneider, T.R, Uson, I, Scheldrick, G.M. | | Deposit date: | 2004-09-14 | | Release date: | 2005-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure of Ecballium Elaterium Trypsin Inhibitor II (Eeti-II): A Rigid Molecular Scaffold
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1W80
 
 | | Crystal structure of the alpha-adaptin appendage domain, from the AP2 adaptor complex, bound to 2 peptides from Synaptojanin170 | | Descriptor: | ADAPTER-RELATED PROTEIN COMPLEX 2 ALPHA 2 SUBUNIT, BENZAMIDINE, CARBONATE ION, ... | | Authors: | Ford, M.G.J, Praefcke, G.J.K, McMahon, H.T. | | Deposit date: | 2004-09-14 | | Release date: | 2004-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolving Nature of the Ap2 Alpha-Appendage Hub During Clathrin-Coated Vesicle Endocytosis.
Embo J., 23, 2004
|
|
1W81
 
 | | Crystal structure of apical membrane antigen 1 from Plasmodium vivax | | Descriptor: | APICAL MEMBRANE ANTIGEN 1, IMIDAZOLE | | Authors: | Pizarro, J.C, Vulliez-Le Normand, B, Chesne-Seck, M.-L, Kocken, C.H.M, Thomas, A.W, Bentley, G.A. | | Deposit date: | 2004-09-15 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of the Malaria Vaccine Candidate Apical Membrane Antigen 1
Science, 308, 2005
|
|
1W82
 
 | | p38 Kinase crystal structure in complex with small molecule inhibitor | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 14, N-[(3Z)-5-TERT-BUTYL-2-PHENYL-1,2-DIHYDRO-3H-PYRAZOL-3-YLIDENE]-N'-(4-CHLOROPHENYL)UREA | | Authors: | Tickle, J, Jhoti, H, Cleasby, A, Devine, L. | | Deposit date: | 2004-09-16 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of Novel P38Alpha Map Kinase Inhibitors Using Fragment-Based Lead Generation
J.Med.Chem., 48, 2005
|
|
1W83
 
 | | p38 Kinase crystal structure in complex with small molecule inhibitor | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 14, N-[4-CHLORO-3-(PYRIDIN-3-YLOXYMETHYL)-PHENYL]-3-FLUORO- | | Authors: | Tickle, J, Jhoti, H, Cleasby, A, Devine, L. | | Deposit date: | 2004-09-16 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of novel p38alpha MAP kinase inhibitors using fragment-based lead generation.
J. Med. Chem., 48, 2005
|
|
1W84
 
 | | p38 Kinase crystal structure in complex with small molecule inhibitor | | Descriptor: | 3-(2-PYRIDIN-4-YLETHYL)-1H-INDOLE, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Tickle, J, Jhoti, H, Cleasby, A, Devine, L. | | Deposit date: | 2004-09-16 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of novel p38alpha MAP kinase inhibitors using fragment-based lead generation.
J. Med. Chem., 48, 2005
|
|
1W85
 
 | | The crystal structure of pyruvate dehydrogenase E1 bound to the peripheral subunit binding domain of E2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE COMPONENT OF PYRUVATE, MAGNESIUM ION, ... | | Authors: | Frank, R.A.W, Pratap, J.V, Pei, X.Y, Perham, R.N, Luisi, B.F. | | Deposit date: | 2004-09-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A molecular switch and proton wire synchronize the active sites in thiamine enzymes.
Science, 306, 2004
|
|
1W86
 
 | |
1W87
 
 | |
1W88
 
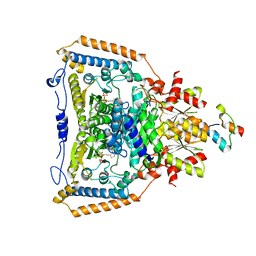 | | The crystal structure of pyruvate dehydrogenase E1(D180N,E183Q) bound to the peripheral subunit binding domain of E2 | | Descriptor: | DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE COMPONENT OF PYRUVATE, MAGNESIUM ION, PYRUVATE DEHYDROGENASE E1 COMPONENT, ... | | Authors: | Frank, R.A.W, Pratap, J.V, Pei, X.Y, Perham, R.N, Luisi, B.F. | | Deposit date: | 2004-09-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Molecular Switch and Proton-Wire Synchronize the Active Sites in Thiamine-Dependent Enzymes
Science, 306, 2004
|
|
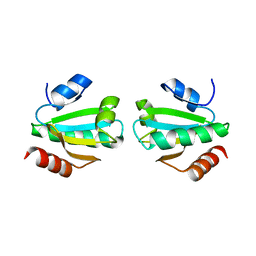
1W89
 
 | |
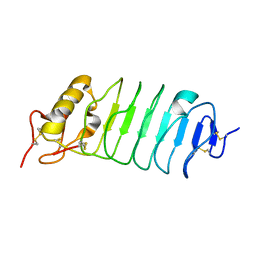
1W8A
 
 | |
1W8B
 
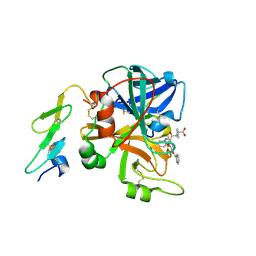 | | Factor7 - 413 complex | | Descriptor: | (S)-[(R)-2-(4-BENZYLOXY-3-METHOXY-PHENYL)-2-(4-CARBAMIMIDOYL-PHENYLAMINO)-ACETYLAMINO]-PHENYL-ACETIC ACID, BLOOD COAGULATION FACTOR VIIA, CALCIUM ION | | Authors: | Ackermann, J, Alig, L, Banner, D.W, Boehm, H.-J, Groebke-Zbinden, K, Hilpert, K, Lave, T, Kuehne, H, Obst-Sander, U, Riederer, M.A, Stahl, M, Tschopp, T.B, Weber, L, Wessel, H.P. | | Deposit date: | 2004-09-17 | | Release date: | 2005-10-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Selective and Orally Bioavailable Phenylglycine Tissue Factor/Factor Viia Inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1W8C
 
 | |
1W8D
 
 | | Binary structure of human DECR. | | Descriptor: | 2,4-DIENOYL-COA REDUCTASE, MITOCHONDRIAL PRECURSOR, GLYCEROL, ... | | Authors: | Alphey, M.S, Byres, E, Hunter, W.N. | | Deposit date: | 2004-09-20 | | Release date: | 2004-10-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Reactivity of Human Mitochondrial 2,4-Dienoyl-Coa Reductase: Enzyme-Ligand Interactions in a Distinctive Short-Chain Reductase Active Site
J.Biol.Chem., 280, 2005
|
|
1W8E
 
 | | 3D structure of CotA incubated with hydrogen peroxide | | Descriptor: | COPPER (II) ION, GLYCEROL, PEROXIDE ION, ... | | Authors: | Bento, I, Martins, L.O, Lopes, G.G, Carrondo, M.A, Lindley, P.F. | | Deposit date: | 2004-09-21 | | Release date: | 2005-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dioxygen Reduction by Multi-Copper Oxidases; a Structural Perspective.
Dalton Trans., 7, 2005
|
|
1W8F
 
 | | PSEUDOMONAS AERUGINOSA LECTIN II (PA-IIL)COMPLEXED WITH LACTO-N-NEO- FUCOPENTAOSE V(LNPFV) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Perret, S, Sabin, C, Dumon, C, Budova, M, Gautier, C, Galanina, O, Ilia, S, Bovin, N, Nicaise, M, Desmadril, M, Gilboa-Garber, N, Wimmerova, M, Mitchell, E.P, Imberty, A. | | Deposit date: | 2004-09-21 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural Basis for the Interaction between Human Milk Oligosaccharides and the Bacterial Lectin Pa-Iil of Pseudomonas Aeruginosa.
Biochem.J., 389, 2005
|
|
1W8G
 
 | | CRYSTAL STRUCTURE OF E. COLI K-12 YGGS | | Descriptor: | HYPOTHETICAL UPF0001 PROTEIN YGGS, ISOCITRIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sulzenbacher, G, Gruez, A, Spinelli, S, Roig-Zamboni, V, Pagot, F, Bignon, C, Vincentelli, R, Cambillau, C. | | Deposit date: | 2004-09-21 | | Release date: | 2006-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of E. Coli K-12 Yggs
To be Published
|
|
1W8H
 
 | | structure of pseudomonas aeruginosa lectin II (PA-IIL)complexed with lewisA trisaccharide | | Descriptor: | CALCIUM ION, GLYCEROL, PSEUDOMONAS AERUGINOSA LECTIN II, ... | | Authors: | Perret, S, Sabin, C, Dumon, C, Budova, M, Gautier, C, Galanina, O, Ilia, S, Bovin, N, Nicaise, M, Desmadril, M, Gilboa-Garber, N, Wimmerova, M, Mitchell, E.P, Imberty, A. | | Deposit date: | 2004-09-21 | | Release date: | 2005-03-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for the Interaction between Human Milk Oligosaccharides and the Bacterial Lectin Pa-Iil of Pseudomonas Aeruginosa.
Biochem.J., 389, 2005
|
|
1W8I
 
 | | The Structure of gene product af1683 from Archaeoglobus fulgidus. | | Descriptor: | PUTATIVE VAPC RIBONUCLEASE AF_1683 | | Authors: | Midwest Center for Structural Genomics (MCSG), Cuff, M.E, Zhang, R, Ginell, S.L, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A. | | Deposit date: | 2004-09-22 | | Release date: | 2004-11-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of Gene Product Af1683 from Archaeoglobus Fulgidus
To be Published
|
|
1W8J
 
 | |
1W8K
 
 | | Crystal structure of apical membrane antigen 1 from Plasmodium vivax | | Descriptor: | APICAL MEMBRANE ANTIGEN 1, IMIDAZOLE | | Authors: | Pizarro, J.C, Vulliez-Le Normand, B, Chesne-Seck, M.-L, Kocken, C.H.M, Thomas, A.W, Bentley, G.A. | | Deposit date: | 2004-09-23 | | Release date: | 2005-03-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Malaria Vaccine Candidate Apical Membrane Antigen 1
Science, 308, 2005
|
|
1W8L
 
 | |
