3BRN
 
 | | Crystal Structure of AM182 Serotonin Complex | | Descriptor: | Lipocalin, SEROTONIN | | Authors: | Mans, B.J, Ribeiro, J.M, Andersen, J.F. | | Deposit date: | 2007-12-21 | | Release date: | 2008-04-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure, function, and evolution of biogenic amine-binding proteins in soft ticks.
J.Biol.Chem., 283, 2008
|
|
5BMV
 
 | | CRYSTAL STRUCTURE OF TUBULIN-STATHMIN-TTL-Vinblastine COMPLEX | | Descriptor: | (2ALPHA,2'BETA,3BETA,4ALPHA,5BETA)-VINCALEUKOBLASTINE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Chen, Q, Zhang, R. | | Deposit date: | 2015-05-23 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into the Pharmacophore of Vinca Domain Inhibitors of Microtubules.
Mol.Pharmacol., 89, 2016
|
|
3JQK
 
 | | Crystal structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from Thermus Theromophilus HB8 (H32 FORM) | | Descriptor: | ACETATE ION, Molybdenum cofactor biosynthesis protein C, PHOSPHATE ION | | Authors: | Kanaujia, S.P, Jeyakanthan, J, Nakagawa, N, Sekar, K, Baba, S, Chen, L, Liu, Z.-J, Wang, B.-C, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-09-07 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of apo and GTP-bound molybdenum cofactor biosynthesis protein MoaC from Thermus thermophilus HB8
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1J4I
 
 | | crystal structure analysis of the FKBP12 complexed with 000308 small molecule | | Descriptor: | 4-METHYL-2-{[4-(TOLUENE-4-SULFONYL)-THIOMORPHOLINE-3-CARBONYL]-AMINO}-PENTANOIC ACID, FKBP12 | | Authors: | Li, P, Ding, Y, Wang, L, Wu, B, Shu, C, Li, S, Shen, B, Rao, Z. | | Deposit date: | 2001-09-30 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and structure-based study of new potential FKBP12 inhibitors.
Biophys.J., 85, 2003
|
|
3J92
 
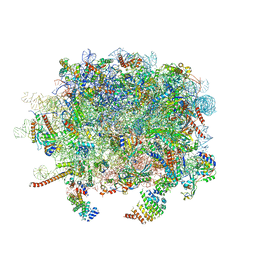 | | Structure and assembly pathway of the ribosome quality control complex | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Shao, S, Brown, A, Santhanam, B, Hegde, R.S. | | Deposit date: | 2014-12-02 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and Assembly Pathway of the Ribosome Quality Control Complex.
Mol.Cell, 57, 2015
|
|
2Q59
 
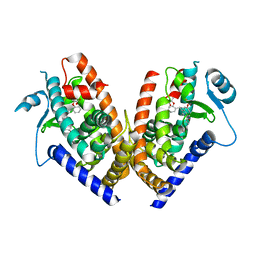 | | Crystal Structure of PPARgamma LBD bound to full agonist MRL20 | | Descriptor: | (2S)-2-(2-{[1-(4-METHOXYBENZOYL)-2-METHYL-5-(TRIFLUOROMETHOXY)-1H-INDOL-3-YL]METHYL}PHENOXY)PROPANOIC ACID, Peroxisome Proliferator-Activated Receptor gamma | | Authors: | Bruning, J.B, Nettles, K.W. | | Deposit date: | 2007-05-31 | | Release date: | 2007-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Partial Agonists Activate PPARgamma Using a Helix 12 Independent Mechanism
Structure, 15, 2007
|
|
1KMB
 
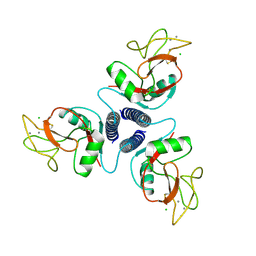 | | SELECTIN-LIKE MUTANT OF MANNOSE-BINDING PROTEIN A | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN-A | | Authors: | Ng, K.K.-S, Weis, W.I. | | Deposit date: | 1996-11-07 | | Release date: | 1997-02-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a selectin-like mutant of mannose-binding protein complexed with sialylated and sulfated Lewis(x) oligosaccharides.
Biochemistry, 36, 1997
|
|
2Q5P
 
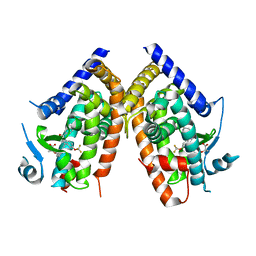 | | Crystal Structure of PPARgamma bound to partial agonist MRL24 | | Descriptor: | (2S)-2-(3-{[1-(4-METHOXYBENZOYL)-2-METHYL-5-(TRIFLUOROMETHOXY)-1H-INDOL-3-YL]METHYL}PHENOXY)PROPANOIC ACID, Peroxisome Proliferator-Activated Receptor gamma | | Authors: | Bruning, J.B, Nettles, K.W. | | Deposit date: | 2007-06-01 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Partial Agonists Activate PPARgamma Using a Helix 12 Independent Mechanism
Structure, 15, 2007
|
|
1FWS
 
 | | AQUIFEX AEOLICUS KDO8P SYNTHASE IN COMPLEX WITH PEP AND CADMIUM | | Descriptor: | 2-DEHYDRO-3-DEOXYPHOSPHOOCTONATE ALDOLASE, CADMIUM ION, PHOSPHATE ION, ... | | Authors: | Duewel, H.S, Radaev, S, Wang, J, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2000-09-24 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Electronic structure of the metal center in the Cd(2+), Zn(2+), and Cu(2+) substituted forms of KDO8P synthase: implications for catalysis.
Biochemistry, 48, 2009
|
|
1EIO
 
 | | ILEAL LIPID BINDING PROTEIN IN COMPLEX WITH GLYCOCHOLATE | | Descriptor: | GLYCOCHOLIC ACID, ILEAL LIPID BINDING PROTEIN | | Authors: | Luecke, C, Zhang, F, Hamilton, J.A, Sacchettini, J.C, Rueterjans, H. | | Deposit date: | 2000-02-27 | | Release date: | 2000-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ileal lipid binding protein in complex with glycocholate.
Eur.J.Biochem., 267, 2000
|
|
2AN5
 
 | | Structure of human PNMT complexed with S-adenosyl-homocysteine and an inhibitor, trans-(1S,2S)-2-amino-1-tetralol | | Descriptor: | PHOSPHATE ION, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Gee, C.L, Tyndall, J.D.A, Grunewald, G.L, Wu, Q, McLeish, M.J, Martin, J.L. | | Deposit date: | 2005-08-11 | | Release date: | 2006-03-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mode of binding of methyl acceptor substrates to the adrenaline-synthesizing enzyme phenylethanolamine N-methyltransferase: implications for catalysis
Biochemistry, 44, 2005
|
|
2MPS
 
 | |
1GAC
 
 | | NMR structure of asymmetric homodimer of a82846b, a glycopeptide antibiotic, complexed with its cell wall pentapeptide fragment | | Descriptor: | CELL WALL PENTAPEPTIDE, CHLOROORIENTICIN A, vancosamine, ... | | Authors: | Kline, A.D, Prowse, W.G, Skelton, M.A, Loncharich, R.J. | | Deposit date: | 1995-05-24 | | Release date: | 1996-08-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Conformation of A82846B, a Glycopeptide Antibiotic, Complexed with its Cell Wall Fragment: An Asymmetric Homodimer Determined Using NMR Spectroscopy.
Biochemistry, 34, 1995
|
|
2AN3
 
 | | Structure of PNMT with S-adenosyl-L-homocysteine and the semi-rigid analogue acceptor substrate cis-(1R,2S)-2-amino-1-tetralol. | | Descriptor: | CIS-(1R,2S)-2-AMINO-1,2,3,4-TETRAHYDRONAPHTHALEN-1-OL, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Gee, C.L, Tyndall, J.D.A, Grunewald, G.L, Wu, Q, McLeish, M.J, Martin, J.L. | | Deposit date: | 2005-08-11 | | Release date: | 2006-03-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mode of binding of methyl acceptor substrates to the adrenaline-synthesizing enzyme phenylethanolamine N-methyltransferase: implications for catalysis
Biochemistry, 44, 2005
|
|
2V7C
 
 | | Crystal Structure of Rev-Erb beta | | Descriptor: | ORPHAN NUCLEAR RECEPTOR NR1D2 | | Authors: | Woo, E.-J, Jeong, D.G, Lim, M.-Y, Kim, S.J, Ryu, S.E. | | Deposit date: | 2007-07-29 | | Release date: | 2007-10-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insight Into the Constitutive Repression Function of the Nuclear Receptor Rev-Erbbeta
J.Mol.Biol., 373, 2007
|
|
1F4X
 
 | |
3OKL
 
 | | Crystal structure of S25-39 in complex with Kdo(2.8)Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-8)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, S25-39 Fab (IgG1k) heavy chain, S25-39 Fab (IgG1k) light chain, ... | | Authors: | Blackler, R.J, Evans, S.V. | | Deposit date: | 2010-08-25 | | Release date: | 2011-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Common NH53K Mutation in the Combining Site of Antibodies Raised against Chlamydial LPS Glycoconjugates Significantly Increases Avidity.
Biochemistry, 50, 2011
|
|
3D0U
 
 | | Crystal Structure of Lysine Riboswitch Bound to Lysine | | Descriptor: | IRIDIUM HEXAMMINE ION, LYSINE, Lysine Riboswitch RNA | | Authors: | Garst, A.D, Heroux, A, Rambo, R.P, Batey, R.T. | | Deposit date: | 2008-05-02 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the lysine riboswitch regulatory mRNA element.
J.Biol.Chem., 283, 2008
|
|
1FWW
 
 | | AQUIFEX AEOLICUS KDO8P SYNTHASE IN COMPLEX WITH PEP, A5P AND CADMIUM | | Descriptor: | 2-DEHYDRO-3-DEOXYPHOSPHOOCTONATE ALDOLASE, ARABINOSE-5-PHOSPHATE, CADMIUM ION, ... | | Authors: | Duewel, H.S, Radaev, S, Wang, J, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2000-09-24 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Electronic structure of the metal center in the Cd(2+), Zn(2+), and Cu(2+) substituted forms of KDO8P synthase: implications for catalysis.
Biochemistry, 48, 2009
|
|
2VDK
 
 | | Re-refinement of Integrin AlphaIIbBeta3 Headpiece | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Springer, T.A, Zhu, J, Xiao, T. | | Deposit date: | 2007-10-10 | | Release date: | 2008-09-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Distinctive Recognition of Fibrinogen Gammac Peptide by the Platelet Integrin Alphaiibbeta3.
J.Cell Biol., 182, 2008
|
|
1GG8
 
 | | DESIGN OF INHIBITORS OF GLYCOGEN PHOSPHORYLASE: A STUDY OF ALPHA-AND BETA-C-GLUCOSIDES AND 1-THIO-BETA-D-GLUCOSE COMPOUNDS | | Descriptor: | ALPHA-D-GLUCOPYRANOSYL-2-CARBOXYLIC ACID AMIDE, INOSINIC ACID, PROTEIN (GLYCOGEN PHOSPHORYLASE), ... | | Authors: | Watson, K.A, Mitchell, E.P, Johnson, L.N, Son, J.C, Bichard, C.J, Orchard, M.G, Fleet, G.W, Oikonomakos, N.G, Leonidas, D.D, Kontou, M, Papageorgiou, A.C. | | Deposit date: | 2000-07-30 | | Release date: | 2000-08-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Design of inhibitors of glycogen phosphorylase: a study of alpha- and beta-C-glucosides and 1-thio-beta-D-glucose compounds.
Biochemistry, 33, 1994
|
|
5JWM
 
 | | Bivalent BET Bromodomain Inhibition | | Descriptor: | 2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]-~{N}-[2-[2-[2-[2-[2-[2-[2-[2-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethyl]ethanamide, Bromodomain-containing protein 4 | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2016-05-12 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Design and characterization of bivalent BET inhibitors.
Nat.Chem.Biol., 12, 2016
|
|
3K90
 
 | | The Abscisic acid receptor PYR1 in complex with Abscisic Acid | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ACETIC ACID, GLYCEROL, ... | | Authors: | Dupeux, F.D, Santiago, J, Rodriguez, P.L, Marquez, J.A. | | Deposit date: | 2009-10-15 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The abscisic acid receptor PYR1 in complex with abscisic acid.
Nature, 462, 2009
|
|
7KJA
 
 | |
5CQT
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | Bromodomain-containing protein 4, N-cyclohexyl-1-ethyl-2-oxo-1,2-dihydrobenzo[cd]indole-6-sulfonamide | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-22 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
