7SXM
 
 | |
8OIK
 
 | | D-PHAT domain (NTD) of human SAMD4A | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Protein Smaug homolog 1 | | Authors: | Kubikova, J, Jeske, M. | | Deposit date: | 2023-03-23 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural basis for binding of Drosophila Smaug to the GPCR Smoothened and to the germline inducer Oskar.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7YRQ
 
 | | Cryo-EM structure of human Peroxisomal ABC Transporter ABCD1 | | Descriptor: | ATP-binding cassette sub-family D member 1, [(2~{R})-1-hexadecanoyloxy-3-[oxidanyl-[2-(trimethyl-$l^{4}-azanyl)ethoxy]phosphoryl]oxy-propan-2-yl] octadec-9-enoate | | Authors: | Chao, X, Li-Na, J, Lin, T. | | Deposit date: | 2022-08-10 | | Release date: | 2023-03-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural insights into substrate recognition and translocation of human peroxisomal ABC transporter ALDP.
Signal Transduct Target Ther, 8, 2023
|
|
6DOM
 
 | |
5HZN
 
 | | Structure of NVP-AEW541 in complex with IGF-1R kinase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7-[cis-3-(azetidin-1-ylmethyl)cyclobutyl]-5-[3-(benzyloxy)phenyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Insulin-like growth factor 1 receptor, ... | | Authors: | Cowan-Jacob, S.W. | | Deposit date: | 2016-02-02 | | Release date: | 2016-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of a 5-[3-phenyl-(2-cyclic-ether)-methylether]-4-aminopyrrolo[2,3-d]pyrimidine series of IGF-1R inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
6DPO
 
 | |
8TWY
 
 | | Structure of p110 alpha bound to (S)-1-(4-((2-(4-(4-(2-amino-4-(difluoromethyl)pyrimidin-5-yl)-6-(3-methylmorpholino)-1,3,5- triazin-2-yl)piperazin-1-yl)-2-oxoethoxy)methyl)piperidin-1-yl)prop-2-en-1-one (compound 9) | | Descriptor: | 1-(4-{[2-(4-{(4P)-4-[2-amino-4-(difluoromethyl)pyrimidin-5-yl]-6-[(3S)-3-methylmorpholin-4-yl]-1,3,5-triazin-2-yl}piperazin-1-yl)-2-oxoethoxy]methyl}piperidin-1-yl)prop-2-en-1-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Burke, J.E, Barlow-Busch, I. | | Deposit date: | 2023-08-21 | | Release date: | 2024-08-28 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Rapid, potent, and persistent covalent chemical probes to deconvolute PI3K alpha signaling.
Chem Sci, 15, 2024
|
|
9LSM
 
 | | The crystal structure of PDE5A with L9 | | Descriptor: | 1-[(13~{S},15~{R})-4-bromanyl-15-(3-chloranyl-4-methoxy-phenyl)-8,12,16-triazatetracyclo[7.7.1.0^{2,7}.0^{13,17}]heptadeca-1(17),2,4,6,8-pentaen-12-yl]ethanone, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Wu, D, Huang, Y.-Y, Luo, H.-B. | | Deposit date: | 2025-02-04 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Plasma metabolites-based drug design: Discovery of novel and highly selective phosphodiesterase 5 inhibitors
Chin.Chem.Lett., 2025
|
|
8U0H
 
 | | Crystal structure of PTPN2 with a PROTAC | | Descriptor: | (5P)-3-(carboxymethoxy)-4-chloro-5-(3-{[(4S)-1-({3-[2-(4-{3-[(3R)-2,6-dioxopiperidin-3-yl]-2-oxo-2,3-dihydro-1,3-benzoxazol-6-yl}piperidin-1-yl)acetamido]phenyl}methanesulfonyl)-2,2-dimethylpiperidin-4-yl]amino}phenyl)thiophene-2-carboxylic acid, ACETATE ION, PTPN2, ... | | Authors: | Jain, R, Longenecker, K, Qiu, W. | | Deposit date: | 2023-08-29 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Mechanistic insights into a heterobifunctional degrader-induced PTPN2/N1 complex.
Commun Chem, 7, 2024
|
|
5HBF
 
 | | Crystal structure of human full-length chitotriosidase (CHIT1) | | Descriptor: | Chitotriosidase-1, GLYCEROL | | Authors: | Fadel, F, Zhao, Y, Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Podjarny, A. | | Deposit date: | 2015-12-31 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-Ray Crystal Structure of the Full Length Human Chitotriosidase (CHIT1) Reveals Features of Its Chitin Binding Domain.
Plos One, 11, 2016
|
|
9N7R
 
 | | Crystal structure of HPK1 bound to N-(3,5-difluoro-4-{[3-(trifluoromethyl)-1H-pyrrolo[2,3-b]pyridin-4-yl]oxy}phenyl)-N'-[3-(morpholin-4-yl)propyl]urea (compound C6) | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Walters, B.T, Kiefer, J.R, Hsu, P.L, Wang, W, Wu, P. | | Deposit date: | 2025-02-06 | | Release date: | 2025-06-25 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Integrating Hydrogen Exchange with Molecular Dynamics for Improved Ligand Binding Predictions.
J.Chem.Inf.Model., 65, 2025
|
|
9NC2
 
 | | Structure of HPK1 with compound C3 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mitogen-activated protein kinase kinase kinase kinase 1, ... | | Authors: | Kiefer, J.R, Walters, B.T, Wang, W, Wu, P, Duo, Y. | | Deposit date: | 2025-02-14 | | Release date: | 2025-06-25 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Integrating Hydrogen Exchange with Molecular Dynamics for Improved Ligand Binding Predictions.
J.Chem.Inf.Model., 65, 2025
|
|
5H6X
 
 | | The crystal structure of RpoS fragment including a partial region 1.2 and region 2 from intracellular pathogen Legionella pneumophila | | Descriptor: | GLYCEROL, RNA polymerase sigma factor RpoS, SODIUM ION | | Authors: | Zhang, N, Chen, X, Gong, X, Ge, H. | | Deposit date: | 2016-11-15 | | Release date: | 2017-11-22 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-Resolution Crystal Structure of RpoS Fragment including a Partial Region 1.2 and Region 2 from the Intracellular Pathogen Legionella pneumophila
Crystals, 8, 2018
|
|
5JN8
 
 | | Crystal Structure for the complex of human carbonic anhydrase IV and acetazolamide | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, ACETATE ION, Carbonic anhydrase 4, ... | | Authors: | Chen, Z, Waheed, A, Di Cera, E, Sly, W.S. | | Deposit date: | 2016-04-29 | | Release date: | 2017-05-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Intrinsic thermodynamics of high affinity inhibitor binding to recombinant human carbonic anhydrase IV.
Eur. Biophys. J., 47, 2018
|
|
9GQV
 
 | |
5EFT
 
 | |
6RU2
 
 | | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-methyl-glucuronoyl methylesterase, ... | | Authors: | Ernst, H.A, Mosbech, C, Langkilde, A, Westh, P, Meyer, A, Agger, J.W, Larsen, S. | | Deposit date: | 2019-05-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The structural basis of fungal glucuronoyl esterase activity on natural substrates.
Nat Commun, 11, 2020
|
|
8I5N
 
 | | Rat Kir4.1 in complex with PIP2 and Lys05 | | Descriptor: | ATP-sensitive inward rectifier potassium channel 10, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Zhao, C, Guo, J. | | Deposit date: | 2023-01-26 | | Release date: | 2024-02-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Pharmacological inhibition of Kir4.1 evokes rapid-onset antidepressant responses.
Nat.Chem.Biol., 20, 2024
|
|
5EHA
 
 | | Crystal structure of recombinant MtaL at 1.35 Angstrom resolution | | Descriptor: | Lectin-like fold protein | | Authors: | Lai, X.-L, Soler-Lopez, M, Wichers, H.J, Dijkstra, B.W. | | Deposit date: | 2015-10-28 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of recombinant tyrosinase-binding protein MtaL at 1.35 angstrom resolution.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5NWX
 
 | | Insight into the molecular recognition mechanism of the coactivator NCoA1 by STAT6 | | Descriptor: | Nuclear receptor coactivator 1, Signal transducer and activator of transcription 6 | | Authors: | Russo, L, Giller, K, Pfitzner, E, Griesinger, C, Becker, S. | | Deposit date: | 2017-05-08 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Insight into the molecular recognition mechanism of the coactivator NCoA1 by STAT6.
Sci Rep, 7, 2017
|
|
7MN9
 
 | | PTP1B 1-284 F225Y-R199N | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Torgeson, K.R, Page, R, Peti, W. | | Deposit date: | 2021-04-30 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Conserved conformational dynamics determine enzyme activity.
Sci Adv, 8, 2022
|
|
7MNA
 
 | | PTP1B 1-284 F225Y-R199N in complex with TCS401 | | Descriptor: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Torgeson, K.R, Page, R, Peti, W. | | Deposit date: | 2021-04-30 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Conserved conformational dynamics determine enzyme activity.
Sci Adv, 8, 2022
|
|
5O3N
 
 | | Crystal structure of E. cloacae 3,4-dihydroxybenzoic acid decarboxylase (AroY) reconstituted with prFMN | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, 3,4-dihydroxybenzoate decarboxylase, GLYCEROL, ... | | Authors: | Marshall, S.A, Leys, D. | | Deposit date: | 2017-05-24 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Regioselective para-Carboxylation of Catechols with a Prenylated Flavin Dependent Decarboxylase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
8IU2
 
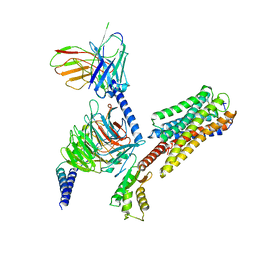 | | Cryo-EM structure of Long-wave-sensitive opsin 1 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Peng, Q, Cheng, X.Y, Li, J, Lu, Q.Y, Li, Y.Y, Zhang, J. | | Deposit date: | 2023-03-23 | | Release date: | 2024-03-27 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM structure of Long-wave-sensitive opsin 1
To Be Published
|
|
5Y0C
 
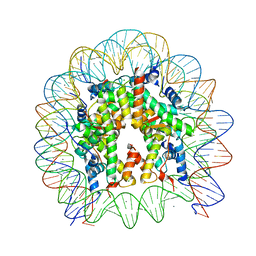 | | Crystal Structure of the human nucleosome at 2.09 angstrom resolution | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Kurumizaka, H, Arimura, Y, Fujita, R, Noda, M. | | Deposit date: | 2017-07-16 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.087 Å) | | Cite: | Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome.
Nucleic Acids Res., 46, 2018
|
|
