6S27
 
 | |
6S4G
 
 | | Crystal structure of the omega transaminase from Chromobacterium violaceum in complex with PMP | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ruggieri, F, Campillo Brocal, J.C, Humble, M.S, Walse, B, Logan, D.T, Berglund, P. | | Deposit date: | 2019-06-27 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Insight into the dimer dissociation process of the Chromobacterium violaceum (S)-selective amine transaminase.
Sci Rep, 9, 2019
|
|
6R75
 
 | | Crystal structure of human Ube2T E54R mutant | | Descriptor: | Ubiquitin-conjugating enzyme E2 T | | Authors: | Chaugule, V.K, Rennie, M.L, Walden, H, Arkinson, C, Kamarainen, O, Toth, R. | | Deposit date: | 2019-03-28 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric mechanism for site-specific ubiquitination of FANCD2.
Nat.Chem.Biol., 16, 2020
|
|
6R89
 
 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with L-cysteine | | Descriptor: | CHLORIDE ION, CYSTEINE, GLYCEROL, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-04-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7D5A
 
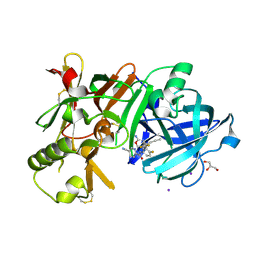 | | Crystal Structure of BACE1 in complex with N-{3-[(9S)-7-amino-2,2-difluoro-9-(prop-1-yn-1-yl)-6-oxa-8-azaspiro[3.5]non-7-en-9-yl]-4-fluorophenyl}-5-cyanopyridine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Fujimoto, K, Yoshida, S, Tadano, G, Asada, N, Fuchino, K, Suzuki, S, Matsuoka, E, Yamamoto, T, Yamamoto, S, Ando, S, Kanegawa, N, Tonomura, Y, Ito, H, Moechars, D, Rombouts, F.J.R, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2020-09-25 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Approaches to Improving Selectivity through Utilizing Explicit Water Molecules: Discovery of Selective beta-Secretase (BACE1) Inhibitors over BACE2.
J.Med.Chem., 64, 2021
|
|
6RBJ
 
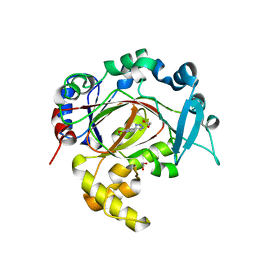 | | Crystal structure of KDM3B in complex with 5-(1H-tetrazol-5-yl)quinolin-8-ol | | Descriptor: | 1,2-ETHANEDIOL, 5-(1~{H}-1,2,3,4-tetrazol-5-yl)quinolin-8-ol, CHLORIDE ION, ... | | Authors: | Johansson, C, Newman, J.A, Kawamura, A, Schofield, C.J, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2019-04-10 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Crystal structure of KDM3B in complex with 5-(1H-tetrazol-5-yl)quinolin-8-ol
To Be Published
|
|
6TOL
 
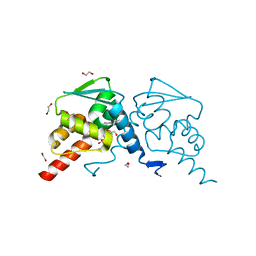 | | Crystal structure of human BCL6 BTB domain in complex with compound 25a | | Descriptor: | 1,2-ETHANEDIOL, 5-[[5-chloranyl-2-[(2~{S},6~{R})-2,6-dimethylmorpholin-4-yl]pyrimidin-4-yl]amino]-1-methyl-3-(3-methyl-3-oxidanyl-butyl)benzimidazol-2-one, B-cell lymphoma 6 protein, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2019-12-11 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | AchievingIn VivoTarget Depletion through the Discovery and Optimization of Benzimidazolone BCL6 Degraders.
J.Med.Chem., 63, 2020
|
|
6TCA
 
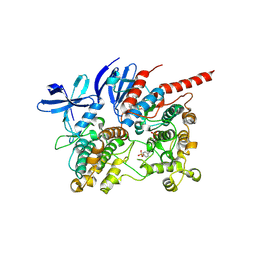 | | Phosphorylated p38 and MAPKAPK2 complex with inhibitor | | Descriptor: | MAP kinase-activated protein kinase 2, Mitogen-activated protein kinase 14, N-[5-(dimethylsulfamoyl)-2-methylphenyl]-1-phenyl-5-propyl-1H-pyrazole-4-carboxamide | | Authors: | Sok, P, Remenyi, A. | | Deposit date: | 2019-11-05 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | MAP Kinase-Mediated Activation of RSK1 and MK2 Substrate Kinases.
Structure, 28, 2020
|
|
6S0H
 
 | | Structure of IMP-13 metallo-beta-lactamase complexed with hydrolysed doripenem | | Descriptor: | (2~{R},3~{R})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-3-methyl-4-[(3~{S},5~{S})-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl]sulfanyl-2,3-dihydro-1~{H}-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Zak, K.M, Softley, C, Kolonko, M, Sattler, M, Popowicz, G.M. | | Deposit date: | 2019-06-14 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure and Molecular Recognition Mechanism of IMP-13 Metallo-beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2020
|
|
6RUQ
 
 | | Structure of GluA2cryst in complex the antagonist ZK200775 and the negative allosteric modulator GYKI53655 at 4.65 A resolution | | Descriptor: | (8R)-5-(4-aminophenyl)-N,8-dimethyl-8,9-dihydro-2H,7H-[1,3]dioxolo[4,5-h][2,3]benzodiazepine-7-carboxamide, Glutamate receptor 2, beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Krintel, C, Venskutonyte, R, Mirza, O.A, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2019-05-28 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.65 Å) | | Cite: | Binding of a negative allosteric modulator and competitive antagonist can occur simultaneously at the ionotropic glutamate receptor GluA2.
Febs J., 288, 2021
|
|
6S43
 
 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate and allosteric modulator N-(5-(Azocan-1-ylsulfonyl)-2-methoxyphenyl)-2-(4-oxo-3,4-dihydrophthalazin-1-yl)acetamide | | Descriptor: | FORMIC ACID, Fumarate hydratase class II, MAGNESIUM ION, ... | | Authors: | Whitehouse, A.J, Libardo, M.D, Kasbekar, M, Brear, P, Fischer, G, Thomas, C.J, Barry, C.E, Boshoff, H.I, Coyne, A.G, Abell, C. | | Deposit date: | 2019-06-26 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Targeting of Fumarate Hydratase fromMycobacterium tuberculosisUsing Allosteric Inhibitors with a Dimeric-Binding Mode.
J.Med.Chem., 62, 2019
|
|
6S54
 
 | | Transaminase from Pseudomonas fluorescens | | Descriptor: | Aspartate aminotransferase family protein, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Smith, P, Roura Padrosa, D, Lopez-Gallego, F, Paradisi, F, Dreveny, I. | | Deposit date: | 2019-06-30 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Enhancing PLP-Binding Capacity of Class-III omega-Transaminase by Single Residue Substitution.
Front Bioeng Biotechnol, 7, 2019
|
|
6S7W
 
 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate and allosteric modulator N-(5-(Azepan-1-ylsulfonyl)-2-methoxyphenyl)-2-(quinolin-4-yl)acetamide | | Descriptor: | FORMIC ACID, Fumarate hydratase class II, MAGNESIUM ION, ... | | Authors: | Whitehouse, A.J, Libardo, M.D, Kasbekar, M, Brear, P, Fischer, G, Thomas, C.J, Barry, C.E, Boshoff, H.I, Coyne, A.G, Abell, C. | | Deposit date: | 2019-07-07 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Targeting of Fumarate Hydratase fromMycobacterium tuberculosisUsing Allosteric Inhibitors with a Dimeric-Binding Mode.
J.Med.Chem., 62, 2019
|
|
6S7K
 
 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate and allosteric modulator N-(2-Methoxy-5-(N-methylsulfamoyl)phenyl)-2-(4-oxo-3,4-dihydrophthalazin-1-yl)acetamide | | Descriptor: | FORMIC ACID, Fumarate hydratase class II, MAGNESIUM ION, ... | | Authors: | Whitehouse, A.J, Libardo, M.D, Kasbekar, M, Brear, P, Fischer, G, Thomas, C.J, Barry, C.E, Boshoff, H.I, Coyne, A.G, Abell, C. | | Deposit date: | 2019-07-05 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Targeting of Fumarate Hydratase fromMycobacterium tuberculosisUsing Allosteric Inhibitors with a Dimeric-Binding Mode.
J.Med.Chem., 62, 2019
|
|
6TT2
 
 | |
6TOJ
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 17a | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[[1-methyl-3-(3-methyl-3-oxidanyl-butyl)-2-oxidanylidene-benzimidazol-5-yl]amino]pyridine-3-carbonitrile, B-cell lymphoma 6 protein | | Authors: | Rodrigues, M.J, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2019-12-11 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | AchievingIn VivoTarget Depletion through the Discovery and Optimization of Benzimidazolone BCL6 Degraders.
J.Med.Chem., 63, 2020
|
|
6TQI
 
 | | I-MOTIF STRUCTURE FORMED FROM THE C STRAND OF A HUMAN TELOMERE FRAGMENT | | Descriptor: | DNA (5'-*TP*AP*AP*CP*CP*CP*TP*AP*A-3') | | Authors: | Parkinson, G.N, Wagner, A, Viladoms-Claverol, J, Duman, R, El-Omari, K. | | Deposit date: | 2019-12-16 | | Release date: | 2020-06-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Native de novo structural determinations of non-canonical nucleic acid motifs by X-ray crystallography at long wavelengths.
Nucleic Acids Res., 48, 2020
|
|
6T37
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Glucose-1-phosphate thymidylyltransferase, ... | | Authors: | Alphey, M.S, Xiao, G, Westwood, J.N. | | Deposit date: | 2019-10-10 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Next generation Glucose-1-phosphate thymidylyltransferase (RmlA) inhibitors: An extended SAR study to direct future design.
Bioorg.Med.Chem., 50, 2021
|
|
6T7L
 
 | | Crystal structure of AmpC from E.coli with Nacubactam (OP0595) | | Descriptor: | (2S,5R)-N-(2-aminoethoxy)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-lactamase, ... | | Authors: | Lang, P.A, Leissing, T.M, Schofield, C.J, Brem, J. | | Deposit date: | 2019-10-22 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Investigations of the Inhibition of Escherichia coli AmpC beta-Lactamase by Diazabicyclooctanes.
Antimicrob.Agents Chemother., 65, 2021
|
|
7G80
 
 | | ARHGEF2 PanDDA analysis group deposition -- ARHGEF2 and RhoA in complex with Z1041785508 | | Descriptor: | 5-(1,3-thiazol-2-yl)-1H-1,2,4-triazole, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-06-22 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.671 Å) | | Cite: | ARHGEF2 PanDDA analysis group deposition
To Be Published
|
|
7G84
 
 | | ARHGEF2 PanDDA analysis group deposition -- ARHGEF2 and RhoA in complex with Z1102357527 | | Descriptor: | DIMETHYL SULFOXIDE, FORMIC ACID, N-[(3R)-6-oxopiperidin-3-yl]-1,3-thiazole-4-carboxamide, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-06-22 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.811 Å) | | Cite: | ARHGEF2 PanDDA analysis group deposition
To Be Published
|
|
7G88
 
 | | ARHGEF2 PanDDA analysis group deposition -- ARHGEF2 and RhoA in complex with Z1148747945 | | Descriptor: | DIMETHYL SULFOXIDE, FORMIC ACID, N-(3-methylpyridin-4-yl)acetamide, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-06-22 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.869 Å) | | Cite: | ARHGEF2 PanDDA analysis group deposition
To Be Published
|
|
7G86
 
 | | ARHGEF2 PanDDA analysis group deposition -- ARHGEF2 and RhoA in complex with Z1137725943 | | Descriptor: | DIMETHYL SULFOXIDE, FORMIC ACID, N-[(1H-indol-4-yl)methyl]ethanamine, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-06-22 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | ARHGEF2 PanDDA analysis group deposition
To Be Published
|
|
7G8B
 
 | | ARHGEF2 PanDDA analysis group deposition -- ARHGEF2 and RhoA in complex with Z100643660 | | Descriptor: | DIMETHYL SULFOXIDE, FORMIC ACID, N,1-dimethyl-1H-indole-3-carboxamide, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-06-22 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | ARHGEF2 PanDDA analysis group deposition
To Be Published
|
|
7G8H
 
 | | ARHGEF2 PanDDA analysis group deposition -- ARHGEF2 and RhoA in complex with Z1198180782 | | Descriptor: | 5-methoxy-1H-pyrrolo[3,2-b]pyridine, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-06-22 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | ARHGEF2 PanDDA analysis group deposition
To Be Published
|
|
