1O50
 
 | |
1O51
 
 | |
1O53
 
 | |
1O54
 
 | |
1O55
 
 | | MOLECULAR STRUCTURE OF TWO CRYSTAL FORMS OF CYCLIC TRIADENYLIC ACID AT 1 ANGSTROM RESOLUTION | | Descriptor: | COBALT (II) ION, DNA (5'-CD(*AP*AP*AP)-3') | | Authors: | Gao, Y.G, Robinson, H, Guan, Y, Liaw, Y.C, van Boom, J.H, van der Marel, G.A, Wang, A.H. | | Deposit date: | 2003-08-20 | | Release date: | 2003-08-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Molecular structure of two crystal forms of cyclic triadenylic acid at 1A resolution.
J.Biomol.Struct.Dyn., 16, 1998
|
|
1O56
 
 | | MOLECULAR STRUCTURE OF TWO CRYSTAL FORMS OF CYCLIC TRIADENYLIC ACID AT 1 ANGSTROM RESOLUTION | | Descriptor: | DNA (5'-CD(*AP*AP*AP*)-3') | | Authors: | Gao, Y.G, Robinson, H, Guan, Y, Liaw, Y.C, van Boom, J.H, van der Marel, G.A, Wang, A.H. | | Deposit date: | 2003-08-20 | | Release date: | 2003-08-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Molecular structure of two crystal forms of cyclic triadenylic acid at 1A resolution.
J.Biomol.Struct.Dyn., 16, 1998
|
|
1O57
 
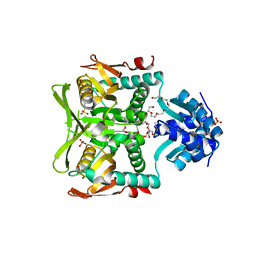 | | CRYSTAL STRUCTURE OF THE PURINE OPERON REPRESSOR OF BACILLUS SUBTILIS | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEXAETHYLENE GLYCOL, NONAETHYLENE GLYCOL, ... | | Authors: | Sinha, S.C, Krahn, J, Shin, B.S, Tomchick, D.R, Zalkin, H, Smith, J.L. | | Deposit date: | 2003-04-20 | | Release date: | 2003-08-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Purine Repressor of Bacillus Subtilis: A Novel Combination of Domains Adapted for Transcription Regulation
J.Bacteriol., 185, 2003
|
|
1O58
 
 | |
1O59
 
 | |
1O5A
 
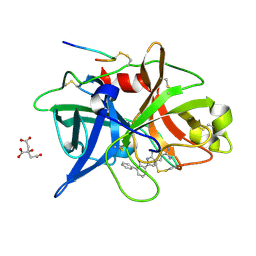 | | Dissecting and Designing Inhibitor Selectivity Determinants at the S1 site Using an Artificial Ala190 Protease (Ala190 uPA) | | Descriptor: | 3-{5-[AMINO(IMINIO)METHYL]-1H-INDOL-2-YL}-1,1'-BIPHENYL-2-OLATE, CITRIC ACID, Urokinase-type plasminogen activator | | Authors: | Katz, B.A, Luong, C, Ho, J.D, Somoza, J.R, Gjerstad, E, Tang, J, Williams, S.R, Verner, E, Mackman, R.L, Young, W.B, Sprengeler, P.A, Chan, H, Mortara, K, Janc, J.W, McGrath, M.E. | | Deposit date: | 2003-09-09 | | Release date: | 2004-09-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Dissecting and designing inhibitor selectivity determinants at the S1 site using an artificial Ala190 protease (Ala190 uPA)
J.Mol.Biol., 344, 2004
|
|
1O5B
 
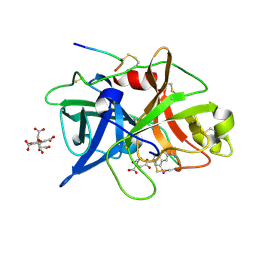 | | Dissecting and Designing Inhibitor Selectivity Determinants at the S1 site Using an Artificial Ala190 Protease (Ala190 uPA) | | Descriptor: | 4-IODOBENZO[B]THIOPHENE-2-CARBOXAMIDINE, CITRIC ACID, Urokinase-type plasminogen activator | | Authors: | Katz, B.A, Luong, C, Ho, J.D, Somoza, J.R, Gjerstad, E, Tang, J, Williams, S.R, Verner, E, Mackman, R.L, Young, W.B, Sprengeler, P.A, Chan, H, Mortara, K, Janc, J.W, McGrath, M.E. | | Deposit date: | 2003-09-09 | | Release date: | 2004-09-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Dissecting and designing inhibitor selectivity determinants at the S1 site using an artificial Ala190 protease (Ala190 uPA)
J.Mol.Biol., 344, 2004
|
|
1O5C
 
 | | Dissecting and Designing Inhibitor Selectivity Determinants at the S1 site Using an Artificial Ala190 Protease (Ala190 uPA) | | Descriptor: | 2-{5-[AMINO(IMINIO)METHYL]-6-FLUORO-1H-BENZIMIDAZOL-2-YL}-6-[(2-METHYLCYCLOHEXYL)OXY]BENZENOLATE, CITRIC ACID, Urokinase-type plasminogen activator | | Authors: | Katz, B.A, Luong, C, Ho, J.D, Somoza, J.R, Gjerstad, E, Tang, J, Williams, S.R, Verner, E, Mackman, R.L, Young, W.B, Sprengeler, P.A, Chan, H, Mortara, K, Janc, J.W, McGrath, M.E. | | Deposit date: | 2003-09-09 | | Release date: | 2004-09-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Dissecting and designing inhibitor selectivity determinants at the S1 site using an artificial Ala190 protease (Ala190 uPA)
J.Mol.Biol., 344, 2004
|
|
1O5D
 
 | | Dissecting and Designing Inhibitor Selectivity Determinants at the S1 site Using an Artificial Ala190 Protease (Ala190 uPA) | | Descriptor: | 2-{5-[AMINO(IMINIO)METHYL]-6-FLUORO-1H-BENZIMIDAZOL-2-YL}-6-[(2-METHYLCYCLOHEXYL)OXY]BENZENOLATE, Coagulation factor VII, Tissue factor | | Authors: | Katz, B.A, Luong, C, Ho, J.D, Somoza, J.R, Gjerstad, E, Tang, J, Williams, S.R, Verner, E, Mackman, R.L, Young, W.B, Sprengeler, P.A, Chan, H, Mortara, K, Janc, J.W, McGrath, M.E. | | Deposit date: | 2003-09-09 | | Release date: | 2004-09-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Dissecting and designing inhibitor selectivity determinants at the S1 site using an artificial Ala190 protease (Ala190 uPA).
J.Mol.Biol., 344, 2004
|
|
1O5E
 
 | | Dissecting and Designing Inhibitor Selectivity Determinants at the S1 site Using an Artificial Ala190 Protease (Ala190 uPA) | | Descriptor: | 6-CHLORO-2-(2-HYDROXY-BIPHENYL-3-YL)-1H-INDOLE-5-CARBOXAMIDINE, Serine protease hepsin | | Authors: | Katz, B.A, Luong, C, Ho, J.D, Somoza, J.R, Gjerstad, E, Tang, J, Williams, S.R, Verner, E, Mackman, R.L, Young, W.B, Sprengeler, P.A, Chan, H, Mortara, K, Janc, J.W, McGrath, M.E. | | Deposit date: | 2003-09-09 | | Release date: | 2004-09-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dissecting and designing inhibitor selectivity determinants at the S1 site using an artificial Ala190 protease (Ala190 uPA).
J.Mol.Biol., 344, 2004
|
|
1O5F
 
 | | Dissecting and Designing Inhibitor Selectivity Determinants at the S1 site Using an Artificial Ala190 Protease (Ala190 uPA) | | Descriptor: | 2-{5-[AMINO(IMINIO)METHYL]-6-FLUORO-1H-BENZIMIDAZOL-2-YL}-6-[(2-METHYLCYCLOHEXYL)OXY]BENZENOLATE, Serine protease hepsin | | Authors: | Katz, B.A, Luong, C, Ho, J.D, Somoza, J.R, Gjerstad, E, Tang, J, Williams, S.R, Verner, E, Mackman, R.L, Young, W.B, Sprengeler, P.A, Chan, H, Mortara, K, Janc, J.W, McGrath, M.E. | | Deposit date: | 2003-09-09 | | Release date: | 2004-09-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Dissecting and designing inhibitor selectivity determinants at the S1 site using an artificial Ala190 protease (Ala190 uPA).
J.Mol.Biol., 344, 2004
|
|
1O5G
 
 | | Dissecting and Designing Inhibitor Selectivity Determinants at the S1 site Using an Artificial Ala190 Protease (Ala190 uPA) | | Descriptor: | 2-{5-[AMINO(IMINIO)METHYL]-6-FLUORO-1H-BENZIMIDAZOL-2-YL}-6-[(2-METHYLCYCLOHEXYL)OXY]BENZENOLATE, CALCIUM ION, Hirudin IIIB', ... | | Authors: | Katz, B.A, Luong, C, Ho, J.D, Somoza, J.R, Gjerstad, E, Tang, J, Williams, S.R, Verner, E, Mackman, R.L, Young, W.B, Sprengeler, P.A, Chan, H, Mortara, K, Janc, J.W, McGrath, M.E. | | Deposit date: | 2003-09-09 | | Release date: | 2004-09-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dissecting and designing inhibitor selectivity determinants at the S1 site using an artificial Ala190 protease (Ala190 uPA).
J.Mol.Biol., 344, 2004
|
|
1O5H
 
 | |
1O5I
 
 | |
1O5J
 
 | |
1O5K
 
 | |
1O5L
 
 | |
1O5M
 
 | | Structure of FPT bound to the inhibitor SCH66336 | | Descriptor: | 4-{2-[4-(3,10-DIBROMO-8-CHLORO-6,11-DIHYDRO-5H-BENZO[5,6]CYCLOHEPTA[1,2-B]PYRIDIN-11-YL)PIPERIDIN-1-YL]-2-OXOETHYL}PIPERIDINE-1-CARBOXAMIDE, FARNESYL DIPHOSPHATE, Protein farnesyltransferase alpha subunit, ... | | Authors: | Strickland, C.L, Weber, P.C, Ganguly, A.K. | | Deposit date: | 2003-09-26 | | Release date: | 2003-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tricyclic Farnesyl Protein Transferase Inhibitors: Crystallographic and Calorimetric Studies of Structure-Activity Relationships
J.Med.Chem., 42, 1999
|
|
1O5O
 
 | |
1O5P
 
 | | Solution Structure of holo-Neocarzinostatin | | Descriptor: | NEOCARZINOSTATIN-CHROMOPHORE, Neocarzinostatin | | Authors: | Takashima, H, Ishino, T, Yoshida, T, Hasuda, K, Ohkubo, T, Kobayashi, Y. | | Deposit date: | 2003-10-04 | | Release date: | 2003-10-14 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure Investigation for Releasing Mechanism of Neocarzinostatin Chromophore from the Holoprotein
J.Biol.Chem., 280, 2005
|
|
1O5Q
 
 | |
