7WR6
 
 | | Crystal structure of ADP-riboxanated caspase-4 in complex with Af1521 | | 分子名称: | ADP-ribose glycohydrolase AF_1521, Caspase-4, [[(3~{a}~{S},5~{R},6~{R},6~{a}~{R})-2-azanylidene-3-[(4~{R})-4-azanyl-5-oxidanylidene-pentyl]-6-oxidanyl-3~{a},5,6,6~{a}-tetrahydrofuro[2,3-d][1,3]oxazol-5-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | 著者 | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | 登録日 | 2022-01-26 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR1
 
 | | P32 of caspase-4 C258A mutant in complex with OspC3 C-terminal ankyrin-repeat domain | | 分子名称: | Caspase-4, OspC3 | | 著者 | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | 登録日 | 2022-01-26 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR3
 
 | | Crystal structure of MBP-fused OspC3 in complex with calmodulin | | 分子名称: | Calmodulin-1, MBP-fused OspC3, NICOTINAMIDE, ... | | 著者 | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | 登録日 | 2022-01-26 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR5
 
 | | Crystal structure of OspC3-calmodulin-caspase-4 complex binding with 2'-aF-NAD+ | | 分子名称: | Calmodulin-1, Caspase-4, OspC3, ... | | 著者 | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | 登録日 | 2022-01-26 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7V6N
 
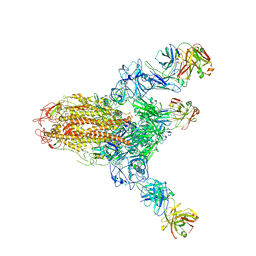 | | MERS S ectodomain trimer in complex with neutralizing antibody 111 state1 | | 分子名称: | 111 H, 111 L, Spike glycoprotein | | 著者 | Wang, X, Zhao, J, Wang, Z, Zeng, J, Zhang, S, Wang, Y. | | 登録日 | 2021-08-20 | | 公開日 | 2022-09-14 | | 実験手法 | ELECTRON MICROSCOPY (3.99 Å) | | 主引用文献 | MERS S ectodomain trimer in complex with neutralizing antibody 111 state1
to be published
|
|
7V6O
 
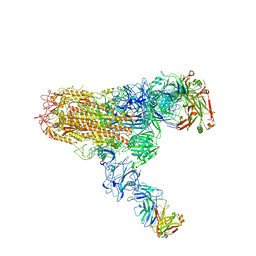 | | MERS S ectodomain trimer in complex with neutralizing antibody 111 (state 2) | | 分子名称: | 111 H, 111 L, Spike glycoprotein | | 著者 | Wang, X, Zhao, J, Wang, Z, Zeng, J, Zhang, S, Wang, Y. | | 登録日 | 2021-08-20 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (4.56 Å) | | 主引用文献 | MERS S ectodomain trimer in complex with neutralizing antibody 111 (state 2)
to be published
|
|
7WQH
 
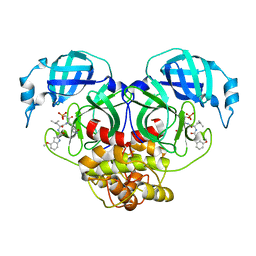 | | Crystal structure of HCoV-NL63 main protease with PF07304814 | | 分子名称: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | 著者 | Zhong, F.L, Zhou, X.L, Lin, C, Zeng, P, Li, J, Zhang, J. | | 登録日 | 2022-01-25 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Structural Basis of Main Proteases of Coronavirus Bound to Drug Candidate PF-07304814
J.Mol.Biol., 434, 2022
|
|
7WQI
 
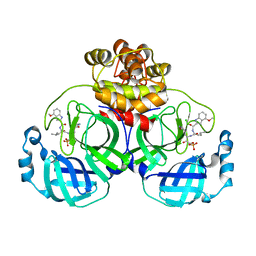 | | Crystal structure of SARS coronavirus main protease in complex with PF07304814 | | 分子名称: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | 著者 | Lin, C, Zhong, F.L, Zhou, X.L, Zeng, P, Zhang, J, Li, J. | | 登録日 | 2022-01-25 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Crystal structure of SARS coronavirus main protease in complex with PF07304814
To Be Published
|
|
7WR4
 
 | | Crystal structure of OspC3-calmodulin-caspase-4 complex | | 分子名称: | Calmodulin-1, Caspase-4, OspC3 | | 著者 | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | 登録日 | 2022-01-26 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3DMV
 
 | |
5XA3
 
 | | Crystal Structure of P450BM3 with Benzyloxycarbonyl-L-prolyl-L-phenylalanine | | 分子名称: | Bifunctional cytochrome P450/NADPH-P450 reductase, DIMETHYL SULFOXIDE, PHENYLALANINE, ... | | 著者 | Shoji, O, Yanagisawa, S, Stanfield, J.K, Suzuki, K, Kasai, C, Cong, Z, Sugimoto, H, Shiro, Y, Watanabe, Y. | | 登録日 | 2017-03-10 | | 公開日 | 2018-02-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Direct Hydroxylation of Benzene to Phenol by Cytochrome P450BM3 Triggered by Amino Acid Derivatives.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
3EYK
 
 | | Structure of Influenza Haemagglutinin in complex with an inhibitor of membrane fusion | | 分子名称: | 2-tert-butylbenzene-1,4-diol, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | 著者 | Russell, R.J, Kerry, P.S, Stevens, D.J, Steinhauer, D.A, Martin, S.R, Gamblin, S.J, Skehel, J.J. | | 登録日 | 2008-10-21 | | 公開日 | 2009-01-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
7D5Z
 
 | | Crystal structure of EBV gH/gL bound with neutralizing antibody 1D8 | | 分子名称: | Envelope glycoprotein H, Envelope glycoprotein L, heavy chain of 1D8, ... | | 著者 | Zhu, Q, Shan, S, Yu, J, Wang, X, Zhang, L, Zeng, M. | | 登録日 | 2020-09-28 | | 公開日 | 2021-10-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (4.2 Å) | | 主引用文献 | A Neutralizing Antibody Targeting a New Site of Vulnerability on Epstein-Barr Virus gH/gL Protects against Dual-Tropic Infection
To Be Published
|
|
7DCB
 
 | |
7DCW
 
 | |
7E8L
 
 | | The structure of Spodoptera litura chemosensory protein | | 分子名称: | Putative chemosensory protein CSP8 | | 著者 | Xie, W, Jia, Q, Zeng, H, Xiao, N, Tang, J, Gao, S, Zhang, J. | | 登録日 | 2021-03-02 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The Crystal Structure of the Spodoptera litura Chemosensory Protein CSP8.
Insects, 12, 2021
|
|
7F79
 
 | | Crystal structure of glutamate dehydrogenase 3 from Candida albicans in complex with alpha-ketoglutarate and NADPH | | 分子名称: | 2-OXOGLUTARIC ACID, GLYCEROL, Glutamate dehydrogenase, ... | | 著者 | Li, N, Wang, W, Zeng, X, Liu, M, Li, M, Li, C, Wang, M. | | 登録日 | 2021-06-28 | | 公開日 | 2021-07-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of glutamate dehydrogenase 3 from Candida albicans.
Biochem.Biophys.Res.Commun., 570, 2021
|
|
7F77
 
 | | Crystal structure of glutamate dehydrogenase 3 from Candida albicans | | 分子名称: | Glutamate dehydrogenase | | 著者 | Li, N, Wang, W, Zeng, X, Liu, M, Li, M, Li, C, Wang, M. | | 登録日 | 2021-06-28 | | 公開日 | 2021-07-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.086 Å) | | 主引用文献 | Crystal structure of glutamate dehydrogenase 3 from Candida albicans.
Biochem.Biophys.Res.Commun., 570, 2021
|
|
7CN8
 
 | | Cryo-EM structure of PCoV_GX spike glycoprotein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein, ... | | 著者 | Wang, X, Yu, J, Zhang, S, Qiao, S, Zeng, J, Tian, L. | | 登録日 | 2020-07-30 | | 公開日 | 2021-03-03 | | 最終更新日 | 2021-03-24 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Bat and pangolin coronavirus spike glycoprotein structures provide insights into SARS-CoV-2 evolution.
Nat Commun, 12, 2021
|
|
7VN0
 
 | | CATPO mutant - T188A | | 分子名称: | ALANINE, CALCIUM ION, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, ... | | 著者 | Yuzugullu Karakus, Y, Balci Unver, S, Zengin Karatas, M, Goc, G, Pearson, A.R, Yorke, B. | | 登録日 | 2021-10-10 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Investigation of how gate residues in the main channel affect the catalytic activity of Scytalidium thermophilum catalase.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7CN4
 
 | | Cryo-EM structure of bat RaTG13 spike glycoprotein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Wang, X, Zhang, S, Qiao, S, Yu, J, Zeng, J, Tian, L. | | 登録日 | 2020-07-30 | | 公開日 | 2021-03-03 | | 最終更新日 | 2021-03-24 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Bat and pangolin coronavirus spike glycoprotein structures provide insights into SARS-CoV-2 evolution.
Nat Commun, 12, 2021
|
|
3EYM
 
 | | Structure of Influenza Haemagglutinin in complex with an inhibitor of membrane fusion | | 分子名称: | 2-tert-butylbenzene-1,4-diol, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | 著者 | Russell, R.J, Kerry, P.S, Stevens, D.A, Steinhauer, D.A, Martin, S.R, Gamblin, S.J, Skehel, J.J. | | 登録日 | 2008-10-21 | | 公開日 | 2009-01-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
7DBF
 
 | |
7DC9
 
 | |
7DCA
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase bound by xanthosine | | 分子名称: | 2,3-dihydroxanthosine, Guanosine deaminase, ZINC ION | | 著者 | Xie, W, Jia, Q, Zeng, H. | | 登録日 | 2020-10-23 | | 公開日 | 2021-09-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The C-terminal loop of Arabidopsis thaliana guanosine deaminase is essential to catalysis.
Chem.Commun.(Camb.), 57, 2021
|
|
