2MWP
 
 | |
6AM5
 
 | |
4XYM
 
 | |
6ALO
 
 | | VioC L-arginine hydroxylase bound to Fe(II), L-arginine, and a peroxysuccinate intermediate | | 分子名称: | 4-peroxy-4-oxobutanoic acid, ARGININE, Alpha-ketoglutarate-dependent L-arginine hydroxylase, ... | | 著者 | Dunham, N.P, Mitchell, A.J, Boal, A.K. | | 登録日 | 2017-08-08 | | 公開日 | 2017-09-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Visualizing the Reaction Cycle in an Iron(II)- and 2-(Oxo)-glutarate-Dependent Hydroxylase.
J. Am. Chem. Soc., 139, 2017
|
|
4Y0Z
 
 | | Trypsin in complex with with BPTI mutant AMINOBUTYRIC ACID | | 分子名称: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | 著者 | Loll, B, Ye, S, Berger, A.A, Muelow, U, Alings, C, Wahl, M.C, Koksch, B. | | 登録日 | 2015-02-06 | | 公開日 | 2015-06-24 | | 最終更新日 | 2018-04-18 | | 実験手法 | X-RAY DIFFRACTION (1.37 Å) | | 主引用文献 | Fluorine teams up with water to restore inhibitor activity to mutant BPTI.
Chem Sci, 6, 2015
|
|
4Y0Q
 
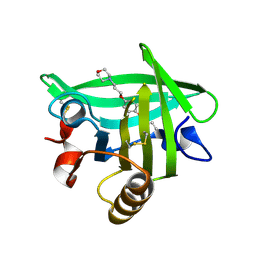 | | Bovine beta-lactoglobulin complex with pramocaine crystallized from sodium citrate (BLG-PRM1) | | 分子名称: | Beta-lactoglobulin, Pramocaine | | 著者 | Loch, J.I, Bonarek, P, Polit, A, Jablonski, M, Czub, M, Ye, X, Lewinski, K. | | 登録日 | 2015-02-06 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | beta-Lactoglobulin interactions with local anaesthetic drugs - Crystallographic and calorimetric studies.
Int.J.Biol.Macromol., 80, 2015
|
|
4Y0V
 
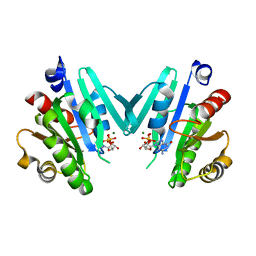 | |
2NB1
 
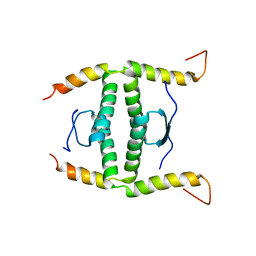 | | P63/p73 hetero-tetramerisation domain | | 分子名称: | Tumor protein 63, Tumor protein p73 | | 著者 | Gebel, J, Buchner, L, Loehr, F.M, Luh, L.M, Coutandin, D, Guentert, P, Doetsch, V. | | 登録日 | 2016-01-19 | | 公開日 | 2016-12-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Mechanism of TAp73 inhibition by Delta Np63 and structural basis of p63/p73 hetero-tetramerization.
Cell Death Differ., 23, 2016
|
|
2MUQ
 
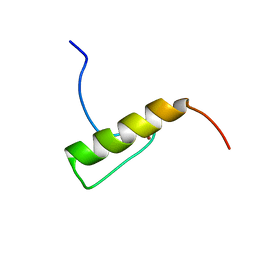 | |
6A16
 
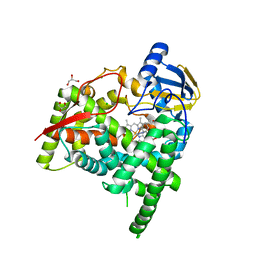 | | Crystal structure of CYP90B1 in complex with uniconazole | | 分子名称: | (1E,3S)-1-(4-chlorophenyl)-4,4-dimethyl-2-(1H-1,2,4-triazol-1-yl)pent-1-en-3-ol, CHLORIDE ION, Cytochrome P450 90B1, ... | | 著者 | Fujiyama, K, Hino, T, Kanadani, M, Mizutani, M, Nagano, S. | | 登録日 | 2018-06-06 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.998 Å) | | 主引用文献 | Structural insights into a key step of brassinosteroid biosynthesis and its inhibition.
Nat.Plants, 5, 2019
|
|
4Y31
 
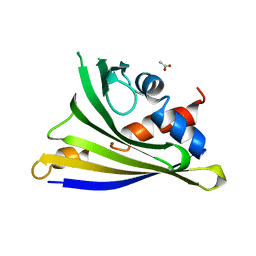 | | Crystal structure of yellow lupine LlPR-10.1A protein in ligand-free form | | 分子名称: | ACETATE ION, Protein LlR18A | | 著者 | Sliwiak, J, Michalska, K, Sikorski, M.M, Jaskolski, M. | | 登録日 | 2015-02-10 | | 公開日 | 2015-12-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Crystallographic and CD probing of ligand-induced conformational changes in a plant PR-10 protein.
J.Struct.Biol., 193, 2016
|
|
5TOJ
 
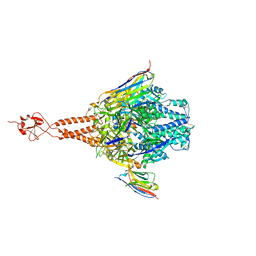 | | Crystal structure of the RSV F glycoprotein in complex with the neutralizing single-domain antibody F-VHH-4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, Fibritin chimera, ... | | 著者 | Gilman, M.S.A, Kabeche, S.C, McLellan, J.S. | | 登録日 | 2016-10-17 | | 公開日 | 2017-02-22 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Potent single-domain antibodies that arrest respiratory syncytial virus fusion protein in its prefusion state.
Nat Commun, 8, 2017
|
|
6A20
 
 | | Crystal Structure of auto-inhibited Kinesin-3 KIF13B | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, HEXAETHYLENE GLYCOL, Kinesin family member 13B, ... | | 著者 | Ren, J.Q, Wang, S, Feng, W. | | 登録日 | 2018-06-08 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Coiled-coil 1-mediated fastening of the neck and motor domains for kinesin-3 autoinhibition.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5TPV
 
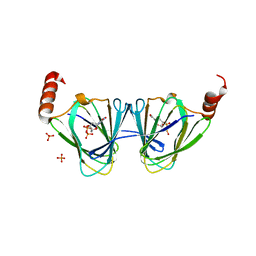 | | X-ray structure of WlaRA (TDP-fucose-3,4-ketoisomerase) from Campylobacter jejuni | | 分子名称: | PHOSPHATE ION, THYMIDINE-5'-DIPHOSPHATE, WlaRA, ... | | 著者 | Holden, H.M, Thoden, J.B, Li, Z.A, Riegert, A.S, Goneau, M.-F, Cunningham, A.M, Vinograd, E, Schoenhofen, I.C, Gilbert, M, Li, J. | | 登録日 | 2016-10-21 | | 公開日 | 2017-02-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Characterization of the dTDP-Fuc3N and dTDP-Qui3N biosynthetic pathways in Campylobacter jejuni 81116.
Glycobiology, 27, 2017
|
|
4Y2Y
 
 | |
2MQW
 
 | |
2MUU
 
 | |
5UHK
 
 | |
2N0J
 
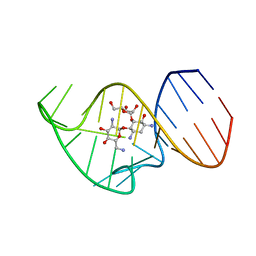 | | Solution NMR Structure of the 27 nucleotide engineered neomycin sensing riboswitch RNA-ribostamycin complex | | 分子名称: | RIBOSTAMYCIN, RNA_(27-MER) | | 著者 | Duchardt-Ferner, E, Gottstein-Schmidtke, S.R, Weigand, J.E, Ohlenschlaeger, O.E, Wurm, J, Hammann, C, Suess, B, Woehnert, J. | | 登録日 | 2015-03-09 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | What a Difference an OH Makes: Conformational Dynamics as the Basis for the Ligand Specificity of the Neomycin-Sensing Riboswitch.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
4XLE
 
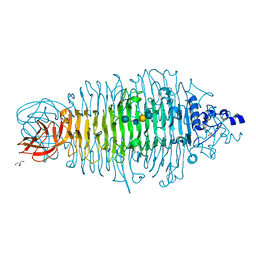 | | Tailspike protein double mutant D339N/E372A of E. coli bacteriophage HK620 in complex with hexasaccharide | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | 著者 | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | 登録日 | 2015-01-13 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
2N2V
 
 | |
5TXN
 
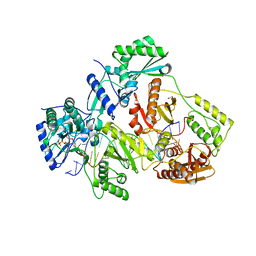 | | STRUCTURE OF Q151M MUTANT HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING DATP | | 分子名称: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3'), ... | | 著者 | Das, K, Martinez, S.M, Arnold, E. | | 登録日 | 2016-11-17 | | 公開日 | 2017-04-05 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structural Insights into HIV Reverse Transcriptase Mutations Q151M and Q151M Complex That Confer Multinucleoside Drug Resistance.
Antimicrob. Agents Chemother., 61, 2017
|
|
4XNH
 
 | |
4XOE
 
 | | Crystal structure of a FimH*DsG complex from E.coli F18 with bound heptyl alpha-D-mannopyrannoside | | 分子名称: | CACODYLATE ION, FimG protein, FimH protein, ... | | 著者 | Jakob, R.P, Sauer, M.M, Navarra, G, Ernst, B, Glockshuber, R, Maier, T. | | 登録日 | 2015-01-16 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Catch-bond mechanism of the bacterial adhesin FimH.
Nat Commun, 7, 2016
|
|
2NO3
 
 | | Novel 4-anilinopyrimidines as potent JNK1 Inhibitors | | 分子名称: | 2-({2-[(3-HYDROXYPHENYL)AMINO]PYRIMIDIN-4-YL}AMINO)BENZAMIDE, C-JUN-AMINO-TERMINAL KINASE-INTERACTING protein 1, Mitogen-activated protein kinase 8, ... | | 著者 | Abad-Zapatero, C. | | 登録日 | 2006-10-24 | | 公開日 | 2007-04-17 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Discovery of a new class of 4-anilinopyrimidines as potent c-Jun N-terminal kinase inhibitors: Synthesis and SAR studies.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
