5DNY
 
 | | Structure of the ATPrS-Mre11/Rad50-DNA complex | | 分子名称: | DNA (27-MER), DNA double-strand break repair Rad50 ATPase,DNA double-strand break repair Rad50 ATPase, DNA double-strand break repair protein Mre11, ... | | 著者 | Liu, Y. | | 登録日 | 2015-09-10 | | 公開日 | 2016-02-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | ATP-dependent DNA binding, unwinding, and resection by the Mre11/Rad50 complex.
Embo J., 35, 2016
|
|
6IID
 
 | | Human EXOG-H140A in complex with RNA-DNA chimeric duplex | | 分子名称: | DNA (5'-D(*CP*GP*TP*GP*AP*CP*AP*TP*CP*CP*CP*G)-3'), DNA/RNA (5'-R(P*CP*GP*GP*GP*A)-D(P*T)-R(P*G)-D(P*T)-R(P*CP*AP*CP*G)-3'), MAGNESIUM ION, ... | | 著者 | Wu, C.C, Lin, J.L.J, Yuan, H.S. | | 登録日 | 2018-10-04 | | 公開日 | 2019-04-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.986 Å) | | 主引用文献 | A unique exonuclease ExoG cleaves between RNA and DNA in mitochondrial DNA replication.
Nucleic Acids Res., 47, 2019
|
|
7YHP
 
 | |
5DNB
 
 | |
7VRU
 
 | | Crystal structure of PacII_M1M2S-DNA-SAH complex | | 分子名称: | DNA (25-mer), S-ADENOSYL-L-HOMOCYSTEINE, Site-specific DNA recognition subunit, ... | | 著者 | Zhu, J, Gao, P. | | 登録日 | 2021-10-25 | | 公開日 | 2022-11-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Molecular insights into DNA recognition and methylation by non-canonical type I restriction-modification systems.
Nat Commun, 13, 2022
|
|
344D
 
 | | DETERMINATION BY MAD-DM OF THE STRUCTURE OF THE DNA DUPLEX D(ACGTACG(5-BRU))2 AT 1.46A AND 100K | | 分子名称: | DNA (5'-D(*AP*CP*GP*TP*AP*CP*GP*(BRU))-3') | | 著者 | Todd, A.R, Adams, A, Powell, H.R, Cardin, C.J. | | 登録日 | 1997-08-04 | | 公開日 | 1997-09-26 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Determination by MAD-DM of the structure of the DNA duplex d[ACGTACG(5-BrU)]2 at 1.46 A and 100 K.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
4BNA
 
 | |
3NM9
 
 | | HMGD(M13A)-DNA complex | | 分子名称: | DNA 5'-D(*G*GP*CP*GP*AP*TP*AP*TP*CP*GP*C)-3', High mobility group protein D | | 著者 | Churchill, M.E.A, Klass, J, Zoetewey, D.L. | | 登録日 | 2010-06-22 | | 公開日 | 2010-09-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structural analysis of HMGD-DNA complexes reveals influence of intercalation on sequence selectivity and DNA bending.
J.Mol.Biol., 403, 2010
|
|
309D
 
 | |
2ORE
 
 | |
2AE9
 
 | | Solution Structure of the theta subunit of DNA polymerase III from E. coli | | 分子名称: | DNA polymerase III, theta subunit | | 著者 | Mueller, G.A, Kirby, T.W, Derose, E.F, Li, D, Schaaper, R.M, London, R.E. | | 登録日 | 2005-07-21 | | 公開日 | 2005-10-18 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Nuclear Magnetic Resonance Solution Structure of the Escherichia coli DNA Polymerase III {theta} Subunit.
J.Bacteriol., 187, 2005
|
|
7W0V
 
 | | C4'-SCF3-DT modifeid DNA-DNA duplex | | 分子名称: | DNA (5'-D(*CP*CP*AP*TP*(DSW)P*AP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*TP*GP*G)-3') | | 著者 | Li, Q, Trajkovski, M, Fan, C, Chen, J, Zhou, Y, Lu, K, Li, H, Su, X, Xi, Z, Plavec, J, Zhou, C. | | 登録日 | 2021-11-18 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | 4'-SCF 3 -Labeling Constitutes a Sensitive 19 F NMR Probe for Characterization of Interactions in the Minor Groove of DNA.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
8SJD
 
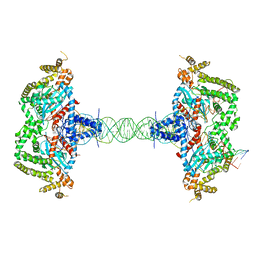 | |
5W2M
 
 | | APOBEC3F Catalytic Domain Complex with a Single-Stranded DNA | | 分子名称: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA dC->dU-editing enzyme APOBEC-3F, ZINC ION | | 著者 | Fang, Y, Xiao, X, Li, S.-X, Wolfe, A, Chen, X.S. | | 登録日 | 2017-06-06 | | 公開日 | 2017-12-13 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Molecular Interactions of a DNA Modifying Enzyme APOBEC3F Catalytic Domain with a Single-Stranded DNA.
J. Mol. Biol., 430, 2018
|
|
1LQG
 
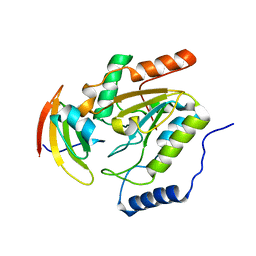 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE COMPLEX WITH URACIL-DNA GLYCOSYLASE INHIBITOR PROTEIN | | 分子名称: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | 著者 | Saikrishnan, K, Sagar, M.B, Ravishankar, R, Roy, S, Purnapatre, K, Handa, P, Varshney, U, Vijayan, M. | | 登録日 | 2002-05-10 | | 公開日 | 2002-11-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Domain closure and action of uracil DNA glycosylase (UDG): structures of new crystal forms containing the Escherichia coli enzyme and a comparative study of the known structures involving UDG.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2Z6U
 
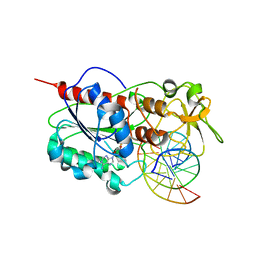 | |
4QEN
 
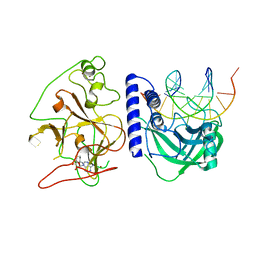 | | crystal structure of KRYPTONITE in complex with mCHH DNA and SAH | | 分子名称: | DNA (5'-D(*AP*CP*TP*GP*AP*TP*GP*AP*GP*TP*AP*CP*CP*AP*T)-3'), DNA (5'-D(*GP*GP*TP*AP*CP*TP*(5CM)P*AP*TP*CP*AP*GP*TP*AP*T)-3'), Histone-lysine N-methyltransferase, ... | | 著者 | Du, J, Li, S, Patel, D.J. | | 登録日 | 2014-05-17 | | 公開日 | 2014-07-30 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.002 Å) | | 主引用文献 | Mechanism of DNA Methylation-Directed Histone Methylation by KRYPTONITE.
Mol.Cell, 55, 2014
|
|
4QEP
 
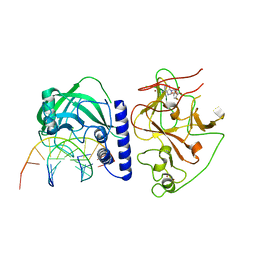 | | crystal structure of KRYPTONITE in complex with mCHG DNA and SAH | | 分子名称: | DNA (5'-D(*AP*CP*TP*GP*CP*TP*GP*AP*GP*TP*AP*CP*CP*AP*T)-3'), DNA (5'-D(*GP*GP*TP*AP*CP*TP*(5CM)P*AP*GP*CP*AP*GP*TP*AP*T)-3'), Histone-lysine N-methyltransferase, ... | | 著者 | Du, J, Li, S, Patel, D.J. | | 登録日 | 2014-05-17 | | 公開日 | 2014-07-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Mechanism of DNA Methylation-Directed Histone Methylation by KRYPTONITE.
Mol.Cell, 55, 2014
|
|
1LQM
 
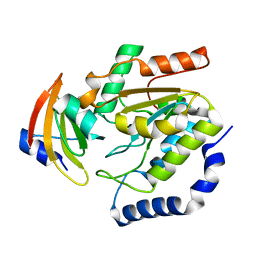 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE COMPLEX WITH URACIL-DNA GLYCOSYLASE INHIBITOR PROTEIN | | 分子名称: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | 著者 | Saikrishnan, K, Sagar, M.B, Ravishankar, R, Roy, S, Purnapatre, K, Varshney, U, Vijayan, M. | | 登録日 | 2002-05-10 | | 公開日 | 2002-11-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Domain closure and action of uracil DNA glycosylase (UDG): structures of new crystal forms containing the Escherichia coli enzyme and a comparative study of the known structures involving UDG.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2LT7
 
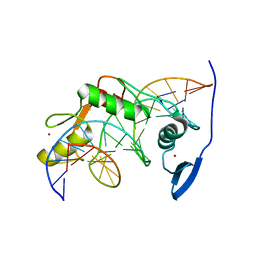 | | Solution NMR structure of Kaiso zinc finger DNA binding domain in complex with Kaiso binding site DNA | | 分子名称: | DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*GP*GP*CP*AP*GP*GP*AP*AP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*TP*TP*CP*CP*TP*GP*CP*CP*AP*AP*TP*AP*AP*CP*G)-3'), Transcriptional regulator Kaiso, ... | | 著者 | Buck-Koehntop, B.A, Stanfield, R.L, Ekiert, D.C, Martinez-Yamout, M.A, Dyson, H, Wilson, I.A, Wright, P.E. | | 登録日 | 2012-05-15 | | 公開日 | 2012-09-05 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
161D
 
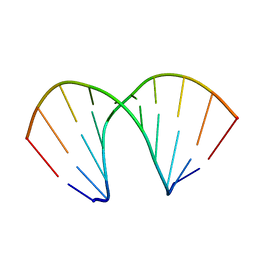 | |
7VS4
 
 | | Crystal structure of PacII_M1M2S-DNA(m6A)-SAH complex | | 分子名称: | DNA (25-mer), S-ADENOSYL-L-HOMOCYSTEINE, Site-specific DNA recognition subunit, ... | | 著者 | Zhu, J, Gao, P. | | 登録日 | 2021-10-25 | | 公開日 | 2022-11-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Molecular insights into DNA recognition and methylation by non-canonical type I restriction-modification systems.
Nat Commun, 13, 2022
|
|
7XQ5
 
 | | Crystal structure of ScIno2p-ScIno4p bound promoter DNA | | 分子名称: | DNA (5'-D(*GP*AP*TP*TP*TP*TP*CP*AP*CP*AP*TP*GP*CP*AP*G)-3'), DNA (5'-D(P*CP*CP*TP*GP*CP*AP*TP*GP*TP*GP*AP*AP*AP*AP*T)-3'), HEXANE-1,6-DIOL, ... | | 著者 | Khan, M.H, Lu, X. | | 登録日 | 2022-05-06 | | 公開日 | 2022-07-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural Analysis of Ino2p/Ino4p Mutual Interactions and Their Binding Interface with Promoter DNA.
Int J Mol Sci, 23, 2022
|
|
4QEO
 
 | | crystal structure of KRYPTONITE in complex with mCHH DNA, H3(1-15) peptide and SAH | | 分子名称: | DNA 5'-ACTGATGAGTACCAT-3', DNA 5'-GGTACT(5CM)ATCAGTAT-3', Histone H3, ... | | 著者 | Du, J, Li, S, Patel, D.J. | | 登録日 | 2014-05-17 | | 公開日 | 2014-07-30 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Mechanism of DNA Methylation-Directed Histone Methylation by KRYPTONITE.
Mol.Cell, 55, 2014
|
|
2AXD
 
 | | solution structure of the theta subunit of escherichia coli DNA polymerase III in complex with the epsilon subunit | | 分子名称: | DNA polymerase III, theta subunit | | 著者 | Keniry, M.A, Park, A.Y, Owen, E.A, Hamdan, S.M, Pintacuda, G, Otting, G, Dixon, N.E. | | 登録日 | 2005-09-05 | | 公開日 | 2006-07-04 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the theta subunit of Escherichia coli DNA polymerase III in complex with the epsilon subunit
J.Bacteriol., 188, 2006
|
|
