2P4S
 
 | | Structure of Purine Nucleoside Phosphorylase from Anopheles gambiae in complex with DADMe-ImmH | | 分子名称: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, Purine nucleoside phosphorylase | | 著者 | Rinaldo-Matthis, A, Almo, S.C, Schramm, V.L. | | 登録日 | 2007-03-13 | | 公開日 | 2008-01-15 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Anopheles gambiae purine nucleoside phosphorylase: catalysis, structure, and inhibition.
Biochemistry, 46, 2007
|
|
1NC1
 
 | | Crystal structure of E. coli MTA/AdoHcy nucleosidase complexed with 5'-methylthiotubercidin (MTH) | | 分子名称: | 2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-METHYLSULFANYLMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, MTA/SAH nucleosidase | | 著者 | Lee, J.E, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | 登録日 | 2002-12-04 | | 公開日 | 2003-11-25 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of Escherichia coli 5'-methylthioadenosine/ S-adenosylhomocysteine nucleosidase inhibitor complexes provide insight into the conformational changes required for substrate binding and catalysis.
J.Biol.Chem., 278, 2003
|
|
3V81
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) with DNA and the nonnucleoside inhibitor nevirapine | | 分子名称: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3'), DNA (5'-D(*AP*TP*GP*GP*AP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), ... | | 著者 | Das, K, Martinez, S.E, Arnold, E. | | 登録日 | 2011-12-22 | | 公開日 | 2012-01-18 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.8503 Å) | | 主引用文献 | HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1NC3
 
 | | Crystal structure of E. coli MTA/AdoHcy nucleosidase complexed with formycin A (FMA) | | 分子名称: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, MTA/SAH nucleosidase | | 著者 | Lee, J.E, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | 登録日 | 2002-12-04 | | 公開日 | 2003-03-25 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of Escherichia coli 5'-methylthioadenosine/ S-adenosylhomocysteine nucleosidase inhibitor complexes provide insight into the conformational changes required for substrate binding and catalysis.
J.Biol.Chem., 278, 2003
|
|
5TUS
 
 | | Potent competitive inhibition of human ribonucleotide reductase by a novel non-nucleoside small molecule | | 分子名称: | 2-hydroxy-N'-[(Z)-(2-hydroxynaphthalen-1-yl)methylidene]benzohydrazide, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large subunit, ... | | 著者 | Mohammed, F.A, Alam, I, Dealwis, C.G. | | 登録日 | 2016-11-07 | | 公開日 | 2017-08-02 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | Potent competitive inhibition of human ribonucleotide reductase by a nonnucleoside small molecule.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3DF9
 
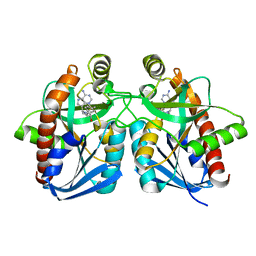 | |
3FZ0
 
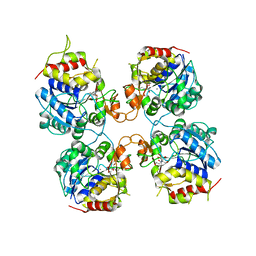 | | Inosine-Guanosine Nucleoside Hydrolase (IG-NH) | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Nucleoside hydrolase, ... | | 著者 | Vandemeulebroucke, A, Minici, C, Bruno, I, Muzzolini, L, Tornaghi, P, Parkin, D.W, Schramm, V.L, Versees, W, Steyaert, J, Degano, M. | | 登録日 | 2009-01-23 | | 公開日 | 2010-01-26 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure and mechanism of the 6-oxopurine nucleosidase from Trypanosoma brucei brucei
Biochemistry, 49, 2010
|
|
6AYQ
 
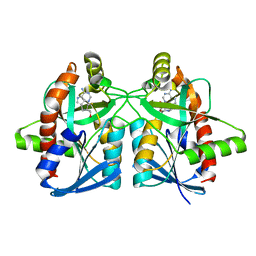 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with methylthio-DADMe-Immucillin-A | | 分子名称: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, GLYCEROL | | 著者 | Cameron, S.A, Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | 登録日 | 2017-09-08 | | 公開日 | 2018-09-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
6AYT
 
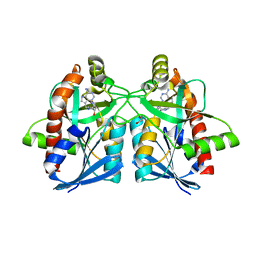 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with pyrazinylthio-DADMe-Immucillin-A | | 分子名称: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(pyrazin-2-ylsulfanyl)methyl]pyrrolidin-3-ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | 著者 | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | 登録日 | 2017-09-08 | | 公開日 | 2018-09-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
6AYO
 
 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with 5'-deoxy-5'-Propyl-DADMe-Immucillin-A | | 分子名称: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-propylpyrrolidin-3-ol, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | 登録日 | 2017-09-08 | | 公開日 | 2018-09-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
6AYR
 
 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with butylthio-DADMe-Immucillin-A | | 分子名称: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(butylsulfanyl)methyl]pyrrolidin-3-ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | 著者 | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | 登録日 | 2017-09-08 | | 公開日 | 2018-09-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
6AYM
 
 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) | | 分子名称: | 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | 著者 | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | 登録日 | 2017-09-08 | | 公開日 | 2018-09-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
6AYS
 
 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with hexylthio-DADMe-Immucillin-A | | 分子名称: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(hexylsulfanyl)methyl]pyrrolidin-3-ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | 著者 | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | 登録日 | 2017-09-08 | | 公開日 | 2018-09-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
1UCN
 
 | | X-ray structure of human nucleoside diphosphate kinase A complexed with ADP at 2 A resolution | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | 著者 | Chen, Y, Gallois-Montbrun, S, Schneider, B, Veron, M, Morera, S, Deville-Bonne, D, Janin, J. | | 登録日 | 2003-04-16 | | 公開日 | 2003-09-30 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Nucleotide Binding to Nucleoside Diphosphate Kinases: X-ray Structure of Human NDPK-A in Complex with ADP and Comparison to Protein Kinases
J.Mol.Biol., 332, 2003
|
|
5TBU
 
 | | Crystal Structure of Isoform 2 of Purine Nucleoside Phosphorylase complexed with Hypoxanthine | | 分子名称: | DIMETHYL SULFOXIDE, HYPOXANTHINE, Purine nucleoside phosphorylase | | 著者 | Faheem, M, Torini, J.R, Romanello, L, Brandao-Neto, J, Pereira, H.M. | | 登録日 | 2016-09-13 | | 公開日 | 2017-10-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The molecular structure of Schistosoma mansoni PNP isoform 2 provides insights into the nucleoside selectivity of PNPs.
PLoS ONE, 13, 2018
|
|
5TBT
 
 | | Crystal Structure of Isoform 2 of Purine Nucleoside Phosphorylase complexed with Cytidine | | 分子名称: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, Purine nucleoside phosphorylase, SULFATE ION | | 著者 | Faheem, M, Torini, J.R, Romanello, L, Brandao-Neto, J, Pereira, H.M. | | 登録日 | 2016-09-13 | | 公開日 | 2017-10-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.101 Å) | | 主引用文献 | The molecular structure of Schistosoma mansoni PNP isoform 2 provides insights into the nucleoside selectivity of PNPs.
PLoS ONE, 13, 2018
|
|
5TBS
 
 | | Crystal Structure of Isoform 2 of Purine Nucleoside Phosphorylase complexed with adenine | | 分子名称: | ADENINE, DIMETHYL SULFOXIDE, Purine nucleoside phosphorylase | | 著者 | Faheem, M, Torini, J.R, Romanello, L, Brandao-Neto, J, Pereira, H.M. | | 登録日 | 2016-09-13 | | 公開日 | 2017-10-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The molecular structure of Schistosoma mansoni PNP isoform 2 provides insights into the nucleoside selectivity of PNPs.
PLoS ONE, 13, 2018
|
|
8RO0
 
 | | Structure of the C. elegans Intron Lariat Spliceosome primed for disassembly (ILS') | | 分子名称: | Cell division cycle 5-like protein, Coiled-coil domain-containing protein 12, GCF C-terminal domain-containing protein, ... | | 著者 | Vorlaender, M.K, Rothe, P, Plaschka, C. | | 登録日 | 2024-01-11 | | 公開日 | 2024-08-07 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Mechanism for the initiation of spliceosome disassembly.
Nature, 632, 2024
|
|
8RO1
 
 | | Structure of the C. elegans Intron Lariat Spliceosome double-primed for disassembly (ILS'') | | 分子名称: | CWF19-like protein 1 homolog, CWF19-like protein 2 homolog, Cell division cycle 5-like protein, ... | | 著者 | Vorlaender, M.K, Rothe, P, Plaschka, C. | | 登録日 | 2024-01-11 | | 公開日 | 2024-08-07 | | 最終更新日 | 2024-08-21 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Mechanism for the initiation of spliceosome disassembly.
Nature, 632, 2024
|
|
3T8I
 
 | | Structural analysis of thermostable S. solfataricus purine-specific nucleoside hydrolase | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Minici, C, Cacciapuoti, G, De Leo, E, Porcelli, M, Degano, M. | | 登録日 | 2011-08-01 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | New Determinants in the Catalytic Mechanism of Nucleoside Hydrolases from the Structures of Two Isozymes from Sulfolobus solfataricus.
Biochemistry, 51, 2012
|
|
2I4T
 
 | | Crystal structure of Purine Nucleoside Phosphorylase from Trichomonas vaginalis with Imm-A | | 分子名称: | 3,4-PYRROLIDINEDIOL,2-(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)-2S,3S,4R,5R, PHOSPHATE ION, Trichomonas vaginalis purine nucleoside phosphorylase | | 著者 | Rinaldo-Matthis, A, Schramm, V.L, Almo, S.C. | | 登録日 | 2006-08-22 | | 公開日 | 2007-06-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.74 Å) | | 主引用文献 | Inhibition and structure of Trichomonas vaginalis purine nucleoside phosphorylase with picomolar transition state analogues.
Biochemistry, 46, 2007
|
|
4I75
 
 | | Crystal structure of the Trypanosoma brucei Inosine-Adenosine-Guanosine nucleoside hydrolase in complex with the NiTris metalorganic complex | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Inosine-adenosine-guanosine-nucleoside hydrolase, ... | | 著者 | Giannese, F, Degano, M. | | 登録日 | 2012-11-30 | | 公開日 | 2013-08-07 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structures of purine nucleosidase from Trypanosoma brucei bound to isozyme-specific trypanocidals and a novel metalorganic inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1Q8F
 
 | |
3T8J
 
 | | Structural analysis of thermostable S. solfataricus pyrimidine-specific nucleoside hydrolase | | 分子名称: | Purine nucleosidase, (IunH-1), SODIUM ION | | 著者 | Minici, C, Cacciapuoti, G, De Leo, E, Porcelli, M, Degano, M. | | 登録日 | 2011-08-01 | | 公開日 | 2012-05-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | New Determinants in the Catalytic Mechanism of Nucleoside Hydrolases from the Structures of Two Isozymes from Sulfolobus solfataricus.
Biochemistry, 51, 2012
|
|
4I74
 
 | | Crystal structure of the Trypanosoma brucei Inosine-Adenosine-Guanosine nucleoside hydrolase in complex with compound UAMC-00312 and allosterically inhibited by a Ni2+ ion | | 分子名称: | (2R,3R,4S)-2-(hydroxymethyl)-1-[(4-hydroxythieno[3,2-d]pyrimidin-7-yl)methyl]pyrrolidine-3,4-diol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | 著者 | Giannese, F, Degano, M. | | 登録日 | 2012-11-30 | | 公開日 | 2013-08-07 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Structures of purine nucleosidase from Trypanosoma brucei bound to isozyme-specific trypanocidals and a novel metalorganic inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
