336D
 
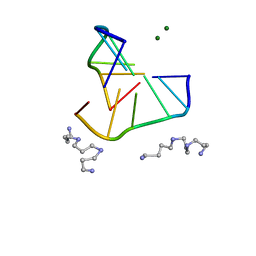 | | INTERACTION BETWEEN LEFT-HANDED Z-DNA AND POLYAMINE-3 THE CRYSTAL STRUCTURE OF THE D(CG)3 AND THERMOSPERMINE COMPLEX | | 分子名称: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), MAGNESIUM ION, N-(3-AMINO-PROPYL)-N-(5-AMINOPROPYL)-1,4-DIAMINOBUTANE | | 著者 | Ohishi, H, Terasoma, N, Nakanishi, I, Van Der Marel, G, Van Boom, J.H, Rich, A, Wang, A.H.-J, Hakoshima, T, Tomita, K.-I. | | 登録日 | 1997-06-24 | | 公開日 | 1998-04-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Interaction between left-handed Z-DNA and polyamine - 3. The crystal structure of the d(CG)3 and thermospermine complex.
FEBS Lett., 398, 1996
|
|
307D
 
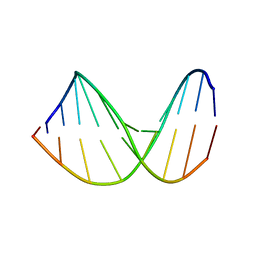 | | Structure of a DNA analog of the primer for HIV-1 RT second strand synthesis | | 分子名称: | DNA (5'-D(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3'), DNA (5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3') | | 著者 | Han, G.W, Kopka, M.L, Cascio, D, Grzeskowiak, K, Dickerson, R.E. | | 登録日 | 1997-01-07 | | 公開日 | 1997-01-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure of a DNA analog of the primer for HIV-1 RT second strand synthesis.
J.Mol.Biol., 269, 1997
|
|
6OZW
 
 | |
2XNK
 
 | |
5MJX
 
 | | 2'F-ANA/DNA Chimeric TBA Quadruplex structure | | 分子名称: | DNA (5'-D(*GP*GP*(FT)P*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3') | | 著者 | Lietard, J, Abou Assi, H, Gomez-Pinto, I, Gonzalez, C, Somoza, M.M, Damha, M.J. | | 登録日 | 2016-12-02 | | 公開日 | 2017-02-01 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Mapping the affinity landscape of Thrombin-binding aptamers on 2 F-ANA/DNA chimeric G-Quadruplex microarrays.
Nucleic Acids Res., 45, 2017
|
|
7KWK
 
 | |
3UXO
 
 | |
2XNH
 
 | |
1D02
 
 | | CRYSTAL STRUCTURE OF MUNI RESTRICTION ENDONUCLEASE IN COMPLEX WITH COGNATE DNA | | 分子名称: | DNA (5'-D(*GP*CP*CP*AP*AP*TP*TP*GP*GP*C)-3'), TYPE II RESTRICTION ENZYME MUNI | | 著者 | Deibert, M, Grazulis, S, Janulaitis, A, Siksnys, V, Huber, R. | | 登録日 | 1999-09-08 | | 公開日 | 2000-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of MunI restriction endonuclease in complex with cognate DNA at 1.7 A resolution.
EMBO J., 18, 1999
|
|
8OLX
 
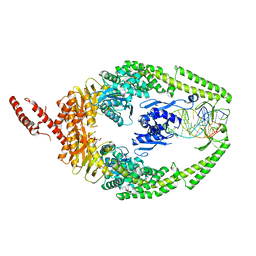 | | MutSbeta bound to (CAG)2 DNA (canonical form) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (25-MER), DNA mismatch repair protein Msh2, ... | | 著者 | Lee, J.-H, Thomsen, M, Daub, H, Steinbacher, S, Sztyler, A, Thieulin-Pardo, G, Neudegger, T, Plotnikov, N, Iyer, R.R, Wilkinson, H, Monteagudo, E, Felsenfeld, D.P, Haque, T, Finley, M, Dominguez, C, Vogt, T.F, Prasad, B.C. | | 登録日 | 2023-03-30 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | MutSbeta bound to (CAG)2 DNA (canonical form)
To Be Published
|
|
7UGW
 
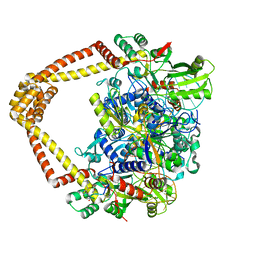 | | M. tuberculosis DNA gyrase cleavage core bound to DNA and evybactin | | 分子名称: | DNA (46-MER), DNA gyrase subunit A, DNA gyrase subunit B, ... | | 著者 | Hauk, G, Imai, Y, Lewis, K, Berger, J.M. | | 登録日 | 2022-03-25 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Evybactin is a DNA gyrase inhibitor that selectively kills Mycobacterium tuberculosis.
Nat.Chem.Biol., 18, 2022
|
|
3UXN
 
 | |
1J8L
 
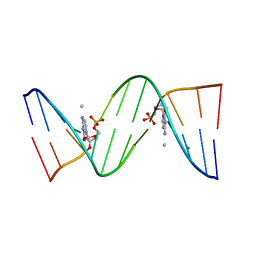 | | Molecular and Crystal Structure of D(CGCAAATTMO4CGCG): the Watson-Crick Type N4-Methoxycytidine/Adenosine Base Pair in B-DNA | | 分子名称: | DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*(C45)P*GP*CP*G)-3'), MAGNESIUM ION | | 著者 | Hossain, M.T, Sunami, T, Tsunoda, M, Hikima, T, Chatake, T, Ueno, Y, Matsuda, A, Takenaka, A. | | 登録日 | 2001-05-22 | | 公開日 | 2001-09-28 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystallographic studies on damaged DNAs IV. N(4)-methoxycytosine shows a second face for Watson-Crick base-pairing, leading to purine transition mutagenesis.
Nucleic Acids Res., 29, 2001
|
|
4HW1
 
 | | Multiple Crystal structures of an all-AT DNA dodecamer stabilized by weak interactions. | | 分子名称: | DNA (5'-D(*AP*AP*TP*AP*AP*AP*TP*TP*TP*AP*TP*T)-3'), MAGNESIUM ION | | 著者 | Acosta-Reyes, F, Subirana, J.A, Pous, J, Condom, N, Malinina, L, Campos, J.L. | | 登録日 | 2012-11-07 | | 公開日 | 2013-11-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Polymorphic crystal structures of an all-AT DNA dodecamer.
Biopolymers, 103, 2015
|
|
1YTF
 
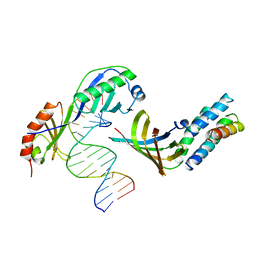 | | YEAST TFIIA/TBP/DNA COMPLEX | | 分子名称: | DNA (5'-D(*GP*TP*TP*TP*TP*AP*TP*AP*TP*AP*CP*AP*TP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*AP*TP*GP*TP*AP*TP*AP*TP*AP*AP*AP*AP*C)-3'), PROTEIN (TATA BINDING PROTEIN (TBP)), ... | | 著者 | Tan, S, Hunziker, Y, Sargent, D.F, Richmond, T.J. | | 登録日 | 1996-04-05 | | 公開日 | 1996-06-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of a yeast TFIIA/TBP/DNA complex.
Nature, 381, 1996
|
|
4R89
 
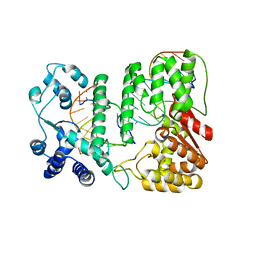 | | Crystal structure of paFAN1 - 5' flap DNA complex with Manganase | | 分子名称: | DNA (5'-D(P*AP*CP*CP*AP*GP*AP*CP*AP*CP*AP*CP*AP*TP*TP*C)-3'), DNA (5'-D(P*GP*AP*AP*TP*GP*TP*GP*TP*GP*TP*CP*TP*CP*AP*AP*TP*CP*CP*CP*AP*AP*C)-3'), DNA (5'-D(P*GP*TP*TP*GP*GP*GP*AP*TP*TP*G)-3'), ... | | 著者 | Cho, Y, Gwon, G.H, Kim, Y.R. | | 登録日 | 2014-08-30 | | 公開日 | 2014-10-29 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (4.002 Å) | | 主引用文献 | Crystal structure of a Fanconi anemia-associated nuclease homolog bound to 5' flap DNA: basis of interstrand cross-link repair by FAN1
Genes Dev., 28, 2014
|
|
383D
 
 | |
1SRS
 
 | |
1D60
 
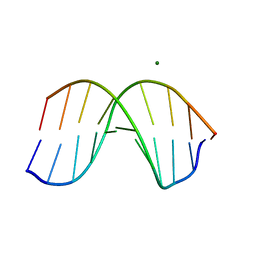 | | THE STRUCTURE OF THE B-DNA DECAMER C-C-A-A-C-I-T-T-G-G: TRIGONAL FORM | | 分子名称: | DNA (5'-D(*CP*CP*AP*AP*CP*IP*TP*TP*GP*G)-3'), MAGNESIUM ION | | 著者 | Lipanov, A, Kopka, M.L, Kaczor-Grzeskowiak, M, Quintana, J, Dickerson, R.E. | | 登録日 | 1992-02-26 | | 公開日 | 1993-04-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of the B-DNA decamer C-C-A-A-C-I-T-T-G-G in two different space groups: conformational flexibility of B-DNA.
Biochemistry, 32, 1993
|
|
2LKX
 
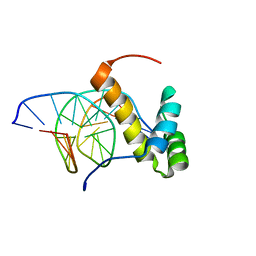 | | NMR structure of the homeodomain of Pitx2 in complex with a TAATCC DNA binding site | | 分子名称: | DNA (5'-D(*CP*GP*GP*GP*GP*AP*TP*TP*AP*GP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*CP*TP*AP*AP*TP*CP*CP*CP*CP*G)-3'), Pituitary homeobox 3 | | 著者 | Baird-Titus, J.M, Doerdelmann, T, Chaney, B.A, Clark-Baldwin, K, Dave, V, Ma, J. | | 登録日 | 2011-10-21 | | 公開日 | 2012-05-02 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the K50 class homeodomain PITX2 bound to DNA and implications for mutations that cause Rieger syndrome
Biochemistry, 44, 2005
|
|
3AV6
 
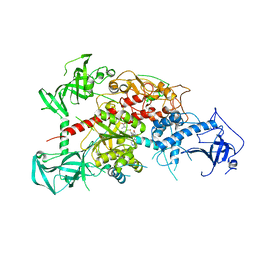 | | Crystal structure of mouse DNA methyltransferase 1 with AdoMet | | 分子名称: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYLMETHIONINE, ZINC ION | | 著者 | Takeshita, K, Suetake, I, Yamashita, E, Suga, M, Narita, H, Nakagawa, A, Tajima, S. | | 登録日 | 2011-02-22 | | 公開日 | 2011-05-04 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3AV5
 
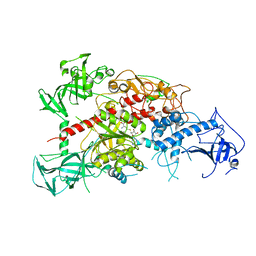 | | Crystal structure of mouse DNA methyltransferase 1 with AdoHcy | | 分子名称: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | 著者 | Takeshita, K, Suetake, I, Yamashita, E, Suga, M, Narita, H, Nakagawa, A, Tajima, S. | | 登録日 | 2011-02-22 | | 公開日 | 2011-05-04 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3S4Z
 
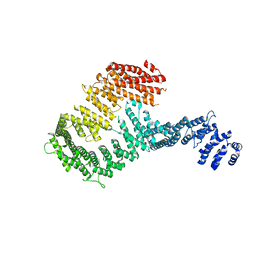 | | Structure of a Y DNA-FANCI complex | | 分子名称: | dna repair 1 | | 著者 | Pavletich, N.P. | | 登録日 | 2011-05-20 | | 公開日 | 2011-07-27 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (7.8 Å) | | 主引用文献 | Structure of the FANCI-FANCD2 complex: insights into the Fanconi anemia DNA repair pathway.
Science, 333, 2011
|
|
8AV6
 
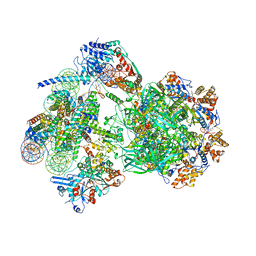 | | CryoEM structure of INO80 core nucleosome complex in closed grappler conformation | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DASH complex subunit DAD4, ... | | 著者 | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | 登録日 | 2022-08-26 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (4.68 Å) | | 主引用文献 | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
8ATF
 
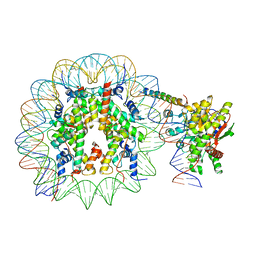 | | Nucleosome-bound Ino80 ATPase | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (226-MER), DNA (227-MER), ... | | 著者 | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | 登録日 | 2022-08-23 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.45 Å) | | 主引用文献 | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
