4BL9
 
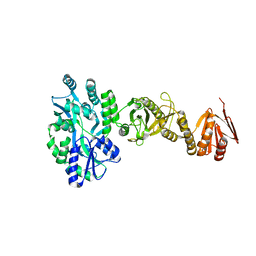 | | Crystal structure of full-length human Suppressor of fused (SUFU) mutant lacking a regulatory subdomain (crystal form I) | | 分子名称: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Cherry, A.L, Finta, C, Karlstrom, M, Toftgard, R, Jovine, L. | | 登録日 | 2013-05-02 | | 公開日 | 2013-11-27 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3PIF
 
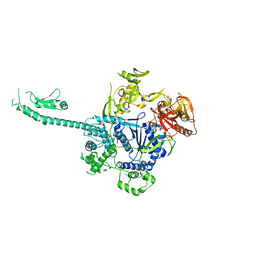 | |
5G4D
 
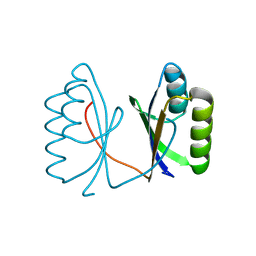 | |
3U8K
 
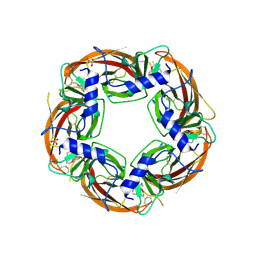 | | Crystal structure of the acetylcholine binding protein (AChBP) from Lymnaea stagnalis in complex with NS3573 (1-(5-ethoxypyridin-3-yl)-1,4-diazepane) | | 分子名称: | 1-(5-ethoxypyridin-3-yl)-1,4-diazepane, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, ... | | 著者 | Rohde, L.A.H, Ahring, P.K, Jensen, M.L, Nielsen, E.O, Peters, D, Helgstrand, C, Krintel, C, Harpsoe, K, Gajhede, M, Kastrup, J.S, Balle, T. | | 登録日 | 2011-10-17 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Intersubunit bridge formation governs agonist efficacy at nicotinic acetylcholine alpha 4 beta 2 receptors: unique role of halogen bonding revealed.
J.Biol.Chem., 287, 2012
|
|
4BLB
 
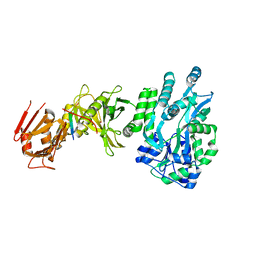 | | Crystal structure of a human Suppressor of fused (SUFU)-GLI1p complex | | 分子名称: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, ZINC FINGER PROTEIN GLI1, ... | | 著者 | Cherry, A.L, Finta, C, Karlstrom, M, De Sanctis, D, Toftgard, R, Jovine, L. | | 登録日 | 2013-05-02 | | 公開日 | 2013-11-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6GIS
 
 | | Structural basis of human clamp sliding on DNA | | 分子名称: | DNA (5'-D(P*AP*TP*AP*CP*GP*AP*TP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*AP*TP*CP*GP*TP*AP*T)-3'), Proliferating cell nuclear antigen | | 著者 | De March, M, Merino, N, Barrera-Vilarmau, S, Crehuet, R, Onesti, S, Blanco, F.J, De Biasio, A. | | 登録日 | 2018-05-15 | | 公開日 | 2018-05-30 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.82 Å) | | 主引用文献 | Structural basis of human PCNA sliding on DNA.
Nat Commun, 8, 2017
|
|
6GMN
 
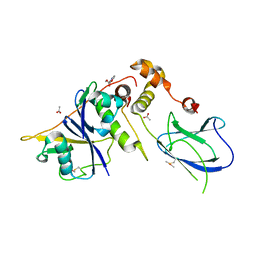 | | pVHL:EloB:EloC in complex with methyl 4H-furo[3,2-b]pyrrole-5-carboxylate | | 分子名称: | ACETATE ION, Elongin-B, Elongin-C, ... | | 著者 | Van Molle, I, Lucas, X, Ciulli, A. | | 登録日 | 2018-05-27 | | 公開日 | 2018-08-08 | | 最終更新日 | 2018-09-05 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Surface Probing by Fragment-Based Screening and Computational Methods Identifies Ligandable Pockets on the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase.
J. Med. Chem., 61, 2018
|
|
5D58
 
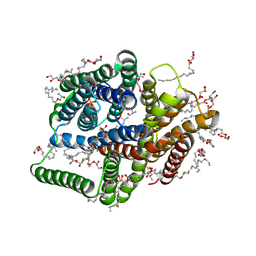 | | In meso in situ serial X-ray crystallography structure of the PepTSt-Ala-Phe complex at 100 K | | 分子名称: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ALANINE, ... | | 著者 | Huang, C.-Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | 登録日 | 2015-08-10 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5D5D
 
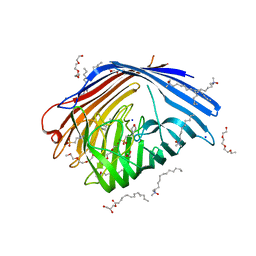 | | In meso in situ serial X-ray crystallography structure of AlgE at 100 K | | 分子名称: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | 著者 | Ma, P, Huang, C.-Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | 登録日 | 2015-08-10 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
3C79
 
 | | Crystal structure of Aplysia californica AChBP in complex with the neonicotinoid imidacloprid | | 分子名称: | (2E)-1-[(6-chloropyridin-3-yl)methyl]-N-nitroimidazolidin-2-imine, ISOPROPYL ALCOHOL, Soluble acetylcholine receptor | | 著者 | Talley, T.T, Harel, M, Hibbs, R.E, Tomizawa, M, Casida, J.E, Taylor, P.W. | | 登録日 | 2008-02-06 | | 公開日 | 2008-05-20 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Atomic interactions of neonicotinoid agonists with AChBP: molecular recognition of the distinctive electronegative pharmacophore.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BLW
 
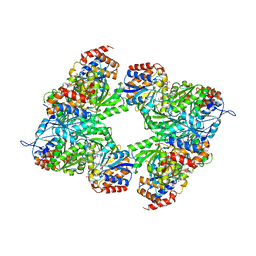 | | Yeast Isocitrate Dehydrogenase with Citrate and AMP Bound in the Regulatory Subunits | | 分子名称: | ADENOSINE MONOPHOSPHATE, CITRATE ANION, Isocitrate dehydrogenase [NAD] subunit 1, ... | | 著者 | Taylor, A.B, Hu, G, Hart, P.J, McAlister-Henn, L. | | 登録日 | 2007-12-11 | | 公開日 | 2008-02-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (4.3 Å) | | 主引用文献 | Allosteric Motions in Structures of Yeast NAD+-specific Isocitrate Dehydrogenase.
J.Biol.Chem., 283, 2008
|
|
3TTB
 
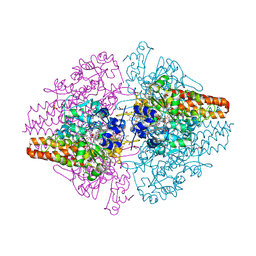 | | Structure of the Thioalkalivibrio paradoxus cytochrome c nitrite reductase in complex with sulfite | | 分子名称: | CALCIUM ION, COBALT (II) ION, Eight-heme nitrite reductase, ... | | 著者 | Polyakov, K.M, Trofimov, A.A, Tikhonova, T.V, Tikhonov, A.V, Dorovatovskii, P.V, Popov, V.O. | | 登録日 | 2011-09-14 | | 公開日 | 2011-10-05 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Comparative structural and functional analysis of two octaheme nitrite reductases from closely related Thioalkalivibrio species.
Febs J., 279, 2012
|
|
3BQ5
 
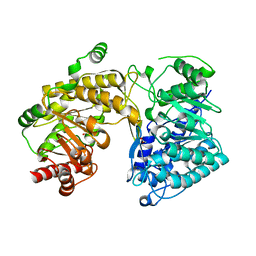 | | Crystal Structure of T. maritima Cobalamin-Independent Methionine Synthase complexed with Zn2+ and Homocysteine (Monoclinic) | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase, ... | | 著者 | Pejchal, R, Smith, J.L, Ludwig, M.L. | | 登録日 | 2007-12-19 | | 公開日 | 2008-03-11 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Metal active site elasticity linked to activation of homocysteine in methionine synthases.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6GZ2
 
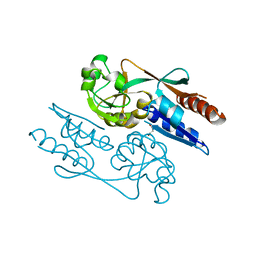 | | Crystal Structure of the LeuO Effector Binding Domain | | 分子名称: | HTH-type transcriptional regulator LeuO, MALONATE ION | | 著者 | Fragel, S, Montada, A.M, Baumann, U, Schacherl, M, Schnetz, K. | | 登録日 | 2018-07-02 | | 公開日 | 2019-06-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Characterization of the pleiotropic LysR-type transcription regulator LeuO of Escherichia coli.
Nucleic Acids Res., 47, 2019
|
|
5WC0
 
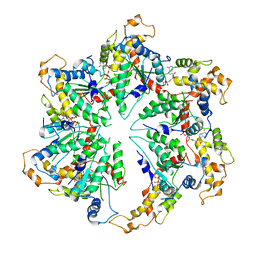 | | katanin hexamer in spiral conformation | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Meiotic spindle formation protein mei-1 | | 著者 | Zehr, E.A, Szyk, A, Piszczek, G, Szczesna, E, Zuo, X, Roll-Mecak, A. | | 登録日 | 2017-06-29 | | 公開日 | 2017-08-09 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Katanin spiral and ring structures shed light on power stroke for microtubule severing.
Nat. Struct. Mol. Biol., 24, 2017
|
|
425D
 
 | | 5'-D(*AP*CP*CP*GP*GP*TP*AP*CP*CP*GP*GP*T)-3' | | 分子名称: | DNA (5'-D(*AP*CP*CP*GP*GP*TP*AP*CP*CP*GP*GP*T)-3') | | 著者 | Rozenberg, H, Rabinovich, D, Frolow, F, Hegde, R.S, Shakked, Z. | | 登録日 | 1998-09-14 | | 公開日 | 1999-10-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural code for DNA recognition revealed in crystal structures of papillomavirus E2-DNA targets.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
233L
 
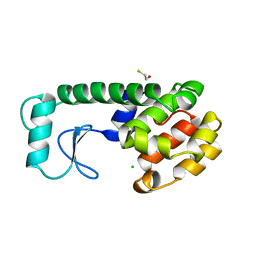 | | T4 LYSOZYME MUTANT M120L | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | 著者 | Lipscomb, L.A, Drew, D.L, Gassner, N, Baase, W.A, Matthews, B.W. | | 登録日 | 1997-10-07 | | 公開日 | 1998-01-14 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Context-dependent protein stabilization by methionine-to-leucine substitution shown in T4 lysozyme.
Protein Sci., 7, 1998
|
|
6H7G
 
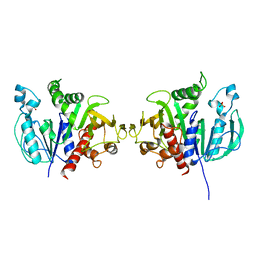 | | Crystal structure of redox-sensitive phosphoribulokinase (PRK) from the green algae Chlamydomonas reinhardtii | | 分子名称: | Phosphoribulokinase, chloroplastic, SULFATE ION | | 著者 | Fermani, S, Sparla, F, Gurrieri, L, Demitri, N, Polentarutti, M, Falini, G, Trost, P, Lemaire, S.D. | | 登録日 | 2018-07-31 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | ArabidopsisandChlamydomonasphosphoribulokinase crystal structures complete the redox structural proteome of the Calvin-Benson cycle.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3BQ4
 
 | |
6GHU
 
 | |
4A7V
 
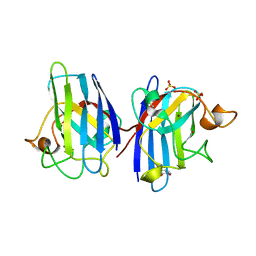 | | Structure of human I113T SOD1 mutant complexed with dopamine in the p21 space group | | 分子名称: | COPPER (II) ION, L-DOPAMINE, SULFATE ION, ... | | 著者 | Wright, G.S.A, Antonyuk, S.V, Kershaw, N.M, Strange, R.W, Hasnain, S.S. | | 登録日 | 2011-11-14 | | 公開日 | 2012-11-28 | | 最終更新日 | 2013-05-08 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Ligand Binding and Aggregation of Pathogenic Sod1.
Nat.Commun., 4, 2013
|
|
4A94
 
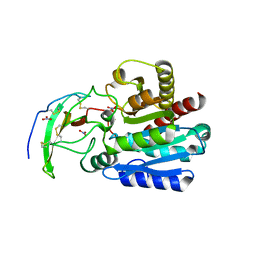 | | Structure of the carboxypeptidase inhibitor from Nerita versicolor in complex with human CPA4 | | 分子名称: | CARBOXYPEPTIDASE A4, CARBOXYPEPTIDASE INHIBITOR, NITRATE ION, ... | | 著者 | Covaleda, G, Alonso, M, Chavez, M.A, Aviles, F.X, Reverter, D. | | 登録日 | 2011-11-23 | | 公開日 | 2011-12-28 | | 最終更新日 | 2012-03-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal Structure of a Novel Metallo-Carboxypeptidase Inhibitor from the Marine Mollusk Nerita Versicolor in Complex with Human Carboxypeptidase A4.
J.Biol.Chem., 287, 2012
|
|
5FJJ
 
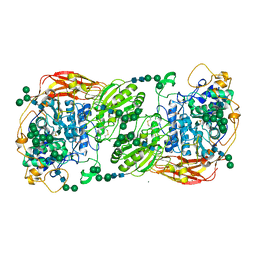 | | Three-dimensional structures of two heavily N-glycosylated Aspergillus sp. Family GH3 beta-D-glucosidases | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-GLUCOSIDASE, ... | | 著者 | Agirre, J, Ariza, A, Offen, W.A, Turkenburg, J.P, Roberts, S.M, McNicholas, S, Harris, P.V, McBrayer, B, Dohnalek, J, Cowtan, K.D, Davies, G.J, Wilson, K.S. | | 登録日 | 2015-10-09 | | 公開日 | 2016-02-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Three-Dimensional Structures of Two Heavily N-Glycosylated Aspergillus Sp. Family Gh3 Beta-D-Glucosidases
Acta Crystallogr.,Sect.D, 72, 2016
|
|
3H1K
 
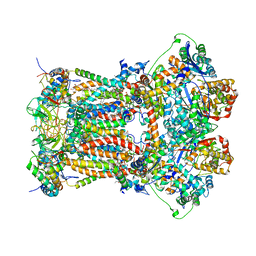 | | Chicken cytochrome BC1 complex with ZN++ and an iodinated derivative of kresoxim-methyl bound | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CARDIOLIPIN, Coenzyme Q10, ... | | 著者 | Berry, E.A, Zhang, Z, Bellamy, H.D, Huang, L.S. | | 登録日 | 2009-04-12 | | 公開日 | 2009-04-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.48 Å) | | 主引用文献 | Crystallographic location of two Zn(2+)-binding sites in the avian cytochrome bc(1) complex
Biochim.Biophys.Acta, 1459, 2000
|
|
6GMR
 
 | | pVHL:EloB:EloC in complex with (4-(1H-pyrrol-1-yl)phenyl)methanol | | 分子名称: | (4-pyrrol-1-ylphenyl)methanol, Elongin-B, Elongin-C, ... | | 著者 | Van Molle, I, Lucas, X, Ciulli, A. | | 登録日 | 2018-05-28 | | 公開日 | 2018-08-08 | | 最終更新日 | 2018-09-05 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Surface Probing by Fragment-Based Screening and Computational Methods Identifies Ligandable Pockets on the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase.
J. Med. Chem., 61, 2018
|
|
