8ZJV
 
 | | Crystal Structure of the ERK2 complexed with 5-Iodotubercidin | | 分子名称: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Yang, Y, Fu, L, Chen, R, Cheng, H, Sun, X. | | 登録日 | 2024-05-15 | | 公開日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of the ERK2 complexed with 5-Iodotubercidin
To Be Published
|
|
8I03
 
 | | Cryo-EM structure of the SIN3L complex from S. pombe | | 分子名称: | Chromatin modification-related protein png2, Histone deacetylase clr6, POTASSIUM ION, ... | | 著者 | Wang, C, Guo, Z, Zhan, X. | | 登録日 | 2023-01-10 | | 公開日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Two assembly modes for SIN3 histone deacetylase complexes.
Cell Discov, 9, 2023
|
|
5M25
 
 | | Modulation of MLL1 Methyltransferase Activity | | 分子名称: | (2~{S})-2-[[(2~{S})-2-[[(2~{S})-2-azanyl-3-oxidanyl-propanoyl]amino]propanoyl]amino]-5-carbamimidamido-~{N}-[(2~{S})-1-[[4-[(~{E})-[4-(hydroxymethyl)phenyl]diazenyl]phenyl]methylamino]-1-oxidanylidene-propan-2-yl]pentanamide, WD repeat-containing protein 5 | | 著者 | Srinivasan, V. | | 登録日 | 2016-10-11 | | 公開日 | 2017-06-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Controlled inhibition of methyltransferases using photoswitchable peptidomimetics: towards an epigenetic regulation of leukemia.
Chem Sci, 8, 2017
|
|
3EUJ
 
 | | Crystal structure of MukE-MukF(residues 292-443)-MukB(head domain)-ATPgammaS complex, symmetric dimer | | 分子名称: | Chromosome partition protein mukB, Linker, Chromosome partition protein mukF, ... | | 著者 | Woo, J.S, Lim, J.H, Shin, H.C, Oh, B.H. | | 登録日 | 2008-10-10 | | 公開日 | 2009-01-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions.
Cell(Cambridge,Mass.), 136, 2009
|
|
3E9G
 
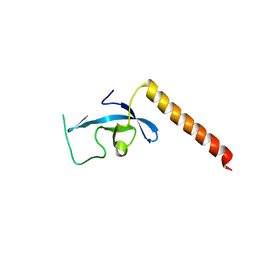 | | Crystal structure long-form (residue1-124) of Eaf3 chromo domain | | 分子名称: | Chromatin modification-related protein EAF3 | | 著者 | Sun, B, Hong, J, Zhang, P, Lin, D, Ding, J. | | 登録日 | 2008-08-22 | | 公開日 | 2008-11-04 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Molecular Basis of the Interaction of Saccharomyces cerevisiae Eaf3 Chromo Domain with Methylated H3K36
J.Biol.Chem., 283, 2008
|
|
7SNZ
 
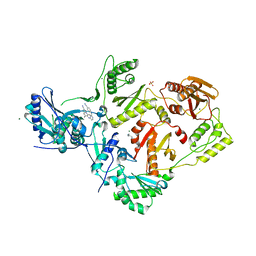 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 6-((2-((4-cyanophenyl)amino)pyrimidin-4-yl)amino)-5,7-dimethylindolizine-2-carbonitrile (JLJ604) | | 分子名称: | (4R)-6-{[2-(4-cyanoanilino)pyrimidin-4-yl]amino}-5,7-dimethylindolizine-2-carbonitrile, 1,4-DIAMINOBUTANE, MAGNESIUM ION, ... | | 著者 | Frey, K.M, Anderson, K.S. | | 登録日 | 2021-10-28 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.368 Å) | | 主引用文献 | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
7SO2
 
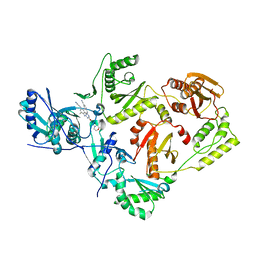 | | Crystal Structure of HIV-1 Reverse Transcriptase Y181C Variant in Complex with (E)-4-((4-((4-(2-cyanovinyl)-2,6-dimethylphenyl)amino)-6-(3-morpholinopropoxy)-1,3,5-triazin-2-yl)amino)benzonitrile (JLJ564) | | 分子名称: | 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylanilino}-6-[3-(morpholin-4-yl)propoxy]-1,3,5-triazin-2-yl)amino]benzonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | 著者 | Frey, K.M, Anderson, K.S. | | 登録日 | 2021-10-29 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.089 Å) | | 主引用文献 | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
7SO6
 
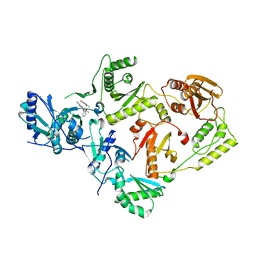 | | Crystal Structure of HIV-1 K103N, Y181C mutant Reverse Transcriptase in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-7-fluoro-2-naphthonitrile (JLJ635), a Non-nucleoside Inhibitor | | 分子名称: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-7-fluoronaphthalene-2-carbonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | 著者 | Bertoletti, N, Frey, K.M, Anderson, K.S, Cisneros Trigo, J.A, Jorgensen, W.L, Chan, A.H. | | 登録日 | 2021-10-29 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
7SNP
 
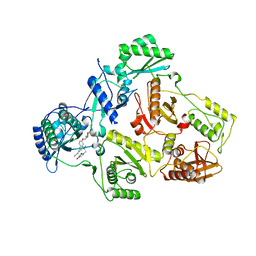 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with (E)-3-(3-chloro-5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)-4-(2-morpholinoethoxy)phenoxy)phenyl)acrylonitrile (JLJ530) | | 分子名称: | (2E)-3-(3-chloro-5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-[2-(morpholin-4-yl)ethoxy]phenoxy}phenyl)prop-2-enenitrile, Reverse transcriptase p66, p51 RT | | 著者 | Frey, K.M, Anderson, K.S. | | 登録日 | 2021-10-28 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
7SO1
 
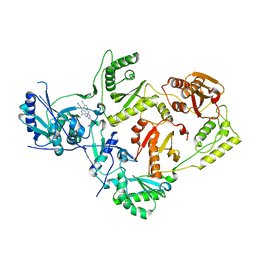 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with (E)-4-((4-((4-(2-cyanovinyl)-2,6-dimethylphenyl)amino)-6-(3-morpholinopropoxy)-1,3,5-triazin-2-yl)amino)benzonitrile (JLJ564) | | 分子名称: | 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylanilino}-6-[3-(morpholin-4-yl)propoxy]-1,3,5-triazin-2-yl)amino]benzonitrile, Reverse transcriptase p66, p51 RT | | 著者 | Frey, K.M, Anderson, K.S. | | 登録日 | 2021-10-28 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.727 Å) | | 主引用文献 | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
7SO4
 
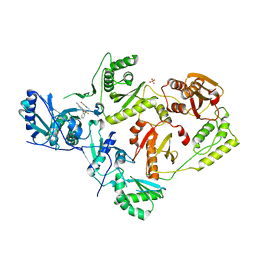 | | Crystal Structure of HIV-1 Y181C mutant Reverse Transcriptase in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-7-fluoro-2-naphthonitrile (JLJ635), a Non-nucleoside Inhibitor | | 分子名称: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-7-fluoronaphthalene-2-carbonitrile, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | 著者 | Bertoletti, N, Anderson, K.S, Cisneros Trigo, J.A, Jorgensen, W.L, Frey, K.M, Chan, A.H. | | 登録日 | 2021-10-29 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
7SO3
 
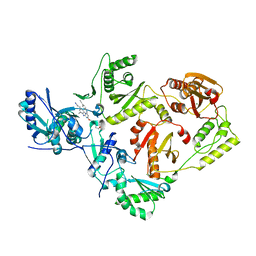 | | Crystal Structure of HIV-1 Reverse Transcriptase K103N/Y181C Variant in Complex with (E)-4-((4-((4-(2-cyanovinyl)-2,6-dimethylphenyl)amino)-6-(3-morpholinopropoxy)-1,3,5-triazin-2-yl)amino)benzonitrile (JLJ564) | | 分子名称: | 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylanilino}-6-[3-(morpholin-4-yl)propoxy]-1,3,5-triazin-2-yl)amino]benzonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | 著者 | Frey, K.M, Anderson, K.S. | | 登録日 | 2021-10-29 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.767 Å) | | 主引用文献 | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
3ERK
 
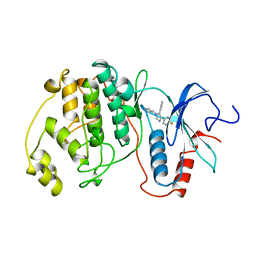 | | THE COMPLEX STRUCTURE OF THE MAP KINASE ERK2/SB220025 | | 分子名称: | 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, EXTRACELLULAR REGULATED KINASE 2 | | 著者 | Wang, Z, Canagarajah, B, Boehm, J.C, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | 登録日 | 1998-07-09 | | 公開日 | 1999-07-22 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
7TDZ
 
 | |
3FK3
 
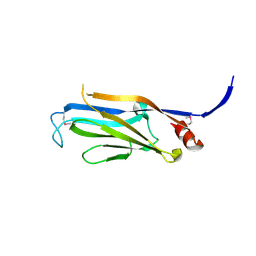 | | Structure of the Yeats Domain, Yaf9 | | 分子名称: | Protein AF-9 homolog | | 著者 | Wang, A.Y, Schulze, J.M, Skordalakes, E, Berger, J.M, Rine, J, Kobor, M.S. | | 登録日 | 2008-12-15 | | 公開日 | 2009-10-27 | | 最終更新日 | 2021-03-31 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Asf1-like structure of the conserved Yaf9 YEATS domain and role in H2A.Z deposition and acetylation
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3F2K
 
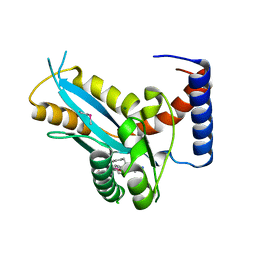 | | Structure of the transposase domain of human Histone-lysine N-methyltransferase SETMAR | | 分子名称: | Histone-lysine N-methyltransferase SETMAR, LYFA Peptide, MAGNESIUM ION | | 著者 | Amaya, M.F, Dombrovski, L, Ni, S, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Botchkarev, A, Min, J, Plotnikov, A.N, Wu, H, Structural Genomics Consortium (SGC) | | 登録日 | 2008-10-29 | | 公開日 | 2008-11-25 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The Crystal Structure of Transposase Domain of Human
Histone-lysine N-methyltransferase SETMAR.
To be Published
|
|
7T3K
 
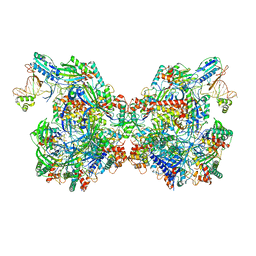 | | Cryo-EM structure of Csy-AcrIF24 dimer | | 分子名称: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR type I-F/YPEST-associated protein Csy3, ... | | 著者 | Mukherjee, I.A, Chang, L. | | 登録日 | 2021-12-08 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural basis of AcrIF24 as an anti-CRISPR protein and transcriptional suppressor.
Nat.Chem.Biol., 18, 2022
|
|
7T3J
 
 | | Cryo-EM structure of Csy-AcrIF24 | | 分子名称: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR type I-F/YPEST-associated protein Csy3, ... | | 著者 | Mukherjee, I.A, Chang, L. | | 登録日 | 2021-12-08 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis of AcrIF24 as an anti-CRISPR protein and transcriptional suppressor.
Nat.Chem.Biol., 18, 2022
|
|
4WQO
 
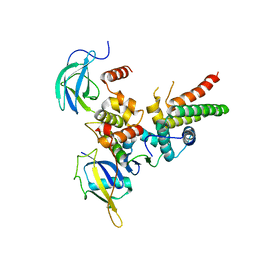 | | Structure of VHL-EloB-EloC-Cul2 | | 分子名称: | Cullin-2, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | 著者 | Nguyen, H.C, Xiong, Y. | | 登録日 | 2014-10-22 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Insights into Cullin-RING E3 Ubiquitin Ligase Recruitment: Structure of the VHL-EloBC-Cul2 Complex.
Structure, 23, 2015
|
|
4UG0
 
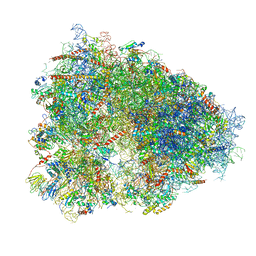 | | STRUCTURE OF THE HUMAN 80S RIBOSOME | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S RIBOSOMAL PROTEIN, ... | | 著者 | Khatter, H, Myasnikov, A.G, Natchiar, S.K, Klaholz, B.P. | | 登録日 | 2015-03-20 | | 公開日 | 2015-06-10 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structure of the human 80S ribosome
NATURE, 520, 2015
|
|
5E81
 
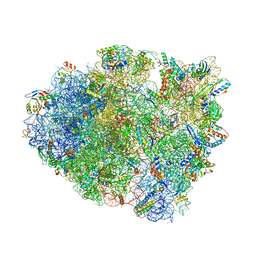 | | Structure of T. thermophilus 70S ribosome complex with mRNA and tRNALys in the A-site with wobble pair | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Rozov, A, Demeshkina, N, Khusainov, I, Yusupov, M, Yusupova, G. | | 登録日 | 2015-10-13 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code.
Nat Commun, 7, 2016
|
|
5EL5
 
 | | Structure of T. thermophilus 70S ribosome complex with mRNA and tRNALys in the A-site with a U-U mismatch in the second position | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Rozov, A, Demeshkina, N, Khusainov, I, Yusupov, M, Yusupova, G. | | 登録日 | 2015-11-04 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code.
Nat Commun, 7, 2016
|
|
5EMN
 
 | | Crystal Structure of Human NADPH-Cytochrome P450 Reductase(A287P mutant) | | 分子名称: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Xia, C, Marohnic, C, Panda, S, Masters, B.S, Kim, J.J.K. | | 登録日 | 2015-11-06 | | 公開日 | 2016-08-17 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Instability of the Human Cytochrome P450 Reductase A287P Variant Is the Major Contributor to Its Antley-Bixler Syndrome-like Phenotype.
J.Biol.Chem., 291, 2016
|
|
5EL6
 
 | | Structure of T. thermophilus 70S ribosome complex with mRNA and tRNALys in the A-site with a U-U mismatch in the first position and antibiotic paromomycin | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Rozov, A, Demeshkina, N, Khusainov, I, Yusupov, M, Yusupova, G. | | 登録日 | 2015-11-04 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code.
Nat Commun, 7, 2016
|
|
5EJB
 
 | | Crystal structure of prefusion Hendra virus F protein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, ... | | 著者 | Wong, J.W, Jardetzky, T.S, Paterson, R.G, Lamb, R.A. | | 登録日 | 2015-11-01 | | 公開日 | 2016-01-06 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structure and stabilization of the Hendra virus F glycoprotein in its prefusion form.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
