1JRW
 
 | | Solution Structure of dAATAA DNA Bulge | | 分子名称: | 5'-D(*CP*GP*TP*AP*GP*CP*CP*GP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*CP*GP*AP*AP*TP*AP*AP*GP*CP*TP*AP*CP*G)-3' | | 著者 | Gollmick, F.A, Lorenz, M, Dornberger, U, von Langen, J, Diekmann, S, Fritzsche, H. | | 登録日 | 2001-08-15 | | 公開日 | 2002-08-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of dAATAA and dAAUAA DNA bulges.
Nucleic Acids Res., 30, 2002
|
|
4UEU
 
 | | Tyrosine kinase AS - a common ancestor of Src and Abl | | 分子名称: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, TYROSINE KINASE AS - A COMMON ANCESTOR OF SRC AND ABL | | 著者 | Kutter, S, Wilson, C, Agafonov, R.V, Hoemberger, M.S, Zorba, A, Halpin, J.C, Theobald, D.L, Kern, D. | | 登録日 | 2014-12-18 | | 公開日 | 2015-02-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Kinase Dynamics. Using Ancient Protein Kinases to Unravel a Modern Cancer Drug'S Mechanism.
Science, 347, 2015
|
|
6JNA
 
 | | Cryo-EM structure of glutamate dehydrogenase from Thermococcus profundus | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate dehydrogenase | | 著者 | Oide, M, Kato, T, Oroguchi, T, Nakasako, M. | | 登録日 | 2019-03-14 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Energy landscape of domain motion in glutamate dehydrogenase deduced from cryo-electron microscopy.
Febs J., 287, 2020
|
|
6JN9
 
 | | Cryo-EM structure of glutamate dehydrogenase from Thermococcus profundus | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate dehydrogenase | | 著者 | Oide, M, Kato, T, Oroguchi, T, Nakasako, M. | | 登録日 | 2019-03-14 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Energy landscape of domain motion in glutamate dehydrogenase deduced from cryo-electron microscopy.
Febs J., 287, 2020
|
|
6JNC
 
 | | Cryo-EM structure of glutamate dehydrogenase from Thermococcus profundus | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate dehydrogenase | | 著者 | Oide, M, Kato, T, Oroguchi, T, Nakasako, M. | | 登録日 | 2019-03-14 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Energy landscape of domain motion in glutamate dehydrogenase deduced from cryo-electron microscopy.
Febs J., 287, 2020
|
|
6JND
 
 | | Cryo-EM structure of glutamate dehydrogenase from Thermococcus profundus | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate dehydrogenase | | 著者 | Oide, M, Kato, T, Oroguchi, T, Nakasako, M. | | 登録日 | 2019-03-14 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Energy landscape of domain motion in glutamate dehydrogenase deduced from cryo-electron microscopy.
Febs J., 287, 2020
|
|
8DZF
 
 | | Cryo-EM structure of bundle-forming pilus extension ATPase from E.coli in the presence of AMP-PNP (class-2) | | 分子名称: | BfpD, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ZINC ION | | 著者 | Nayak, A.R, Zhao, J, Donnenberg, M.S, Samso, M. | | 登録日 | 2022-08-07 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.69 Å) | | 主引用文献 | Cryo-EM Structure of the Type IV Pilus Extension ATPase from Enteropathogenic Escherichia coli.
Mbio, 13, 2022
|
|
8DZE
 
 | | Cryo-EM structure of bundle-forming pilus extension ATPase from E. coli in the presence of AMP-PNP (class-1) | | 分子名称: | BfpD, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ZINC ION | | 著者 | Nayak, A.R, Zhao, J, Donnenberg, M.S, Samso, M. | | 登録日 | 2022-08-07 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | Cryo-EM Structure of the Type IV Pilus Extension ATPase from Enteropathogenic Escherichia coli.
Mbio, 13, 2022
|
|
8DZG
 
 | | Cryo-EM structure of bundle-forming pilus extension ATPase from E.coli in the presence of ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BfpD, MAGNESIUM ION, ... | | 著者 | Nayak, A.R, Zhao, J, Donnenberg, M.S, Samso, M. | | 登録日 | 2022-08-07 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Cryo-EM Structure of the Type IV Pilus Extension ATPase from Enteropathogenic Escherichia coli.
Mbio, 13, 2022
|
|
2KEZ
 
 | | NMR structure of U6 ISL at pH 8.0 | | 分子名称: | RNA (5'-R(*GP*GP*UP*UP*CP*CP*CP*CP*UP*GP*CP*AP*UP*AP*AP*GP*GP*AP*UP*GP*AP*AP*CP*C)-3') | | 著者 | Venditti, V, Butcher, S.E. | | 登録日 | 2009-02-08 | | 公開日 | 2009-07-21 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Minimum-energy path for a u6 RNA conformational change involving protonation, base-pair rearrangement and base flipping.
J.Mol.Biol., 391, 2009
|
|
1YUG
 
 | |
1YUF
 
 | |
2KF0
 
 | | NMR structure of U6 ISL at pH 7.0 | | 分子名称: | RNA (5'-R(*GP*GP*UP*UP*CP*CP*CP*CP*UP*GP*CP*AP*UP*AP*AP*GP*GP*AP*UP*GP*AP*AP*CP*C)-3') | | 著者 | Venditti, V, Butcher, S.E. | | 登録日 | 2009-02-08 | | 公開日 | 2009-07-21 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Minimum-energy path for a u6 RNA conformational change involving protonation, base-pair rearrangement and base flipping.
J.Mol.Biol., 391, 2009
|
|
5EHB
 
 | | A de novo designed hexameric coiled-coil peptide with iodotyrosine | | 分子名称: | pHiosYI | | 著者 | Lizatovic, R, Aurelius, O, Stenstrom, O, Drakenberg, T, Akke, M, Logan, D.T, Andre, I. | | 登録日 | 2015-10-28 | | 公開日 | 2016-06-15 | | 最終更新日 | 2018-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.19 Å) | | 主引用文献 | A De Novo Designed Coiled-Coil Peptide with a Reversible pH-Induced Oligomerization Switch.
Structure, 24, 2016
|
|
1KBH
 
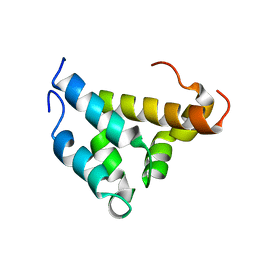 | | Mutual Synergistic Folding in the Interaction Between Nuclear Receptor Coactivators CBP and ACTR | | 分子名称: | CREB-BINDING PROTEIN, nuclear receptor coactivator | | 著者 | Demarest, S.J, Martinez-Yamout, M, Chung, J, Chen, H, Xu, W, Dyson, H.J, Evans, R.M, Wright, P.E. | | 登録日 | 2001-11-06 | | 公開日 | 2002-02-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators.
Nature, 415, 2002
|
|
8CE0
 
 | | N-terminal domain of human apolipoprotein E | | 分子名称: | GLYCEROL, Maltodextrin-binding protein,Apolipoprotein E | | 著者 | Marek, M, Nemergut, M. | | 登録日 | 2023-02-01 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Domino-like effect of C112R mutation on ApoE4 aggregation and its reduction by Alzheimer's Disease drug candidate.
Mol Neurodegener, 18, 2023
|
|
8CDY
 
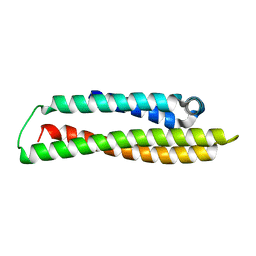 | | N-terminal domain of human apolipoprotein E | | 分子名称: | Maltodextrin-binding protein,Apolipoprotein E | | 著者 | Marek, M, Nemergut, M. | | 登録日 | 2023-02-01 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Domino-like effect of C112R mutation on ApoE4 aggregation and its reduction by Alzheimer's Disease drug candidate.
Mol Neurodegener, 18, 2023
|
|
4WBB
 
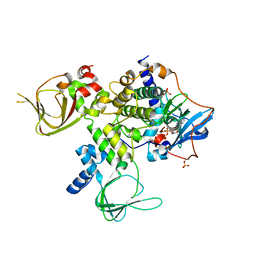 | | Single Turnover Autophosphorylation Cycle of the PKA RIIb Holoenzyme | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | 著者 | Zhang, P, Knape, M.J, Ahuja, L.G, Keshwani, M.M, King, C.C, Sastri, M, Herberg, F.W, Taylor, S.S. | | 登録日 | 2014-09-02 | | 公開日 | 2015-05-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Single Turnover Autophosphorylation Cycle of the PKA RII beta Holoenzyme.
Plos Biol., 13, 2015
|
|
5A5B
 
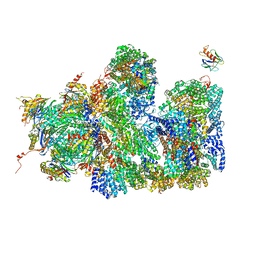 | | Structure of the 26S proteasome-Ubp6 complex | | 分子名称: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASE REGULATORY SUBUNIT 6A, 26S PROTEASE REGULATORY SUBUNIT 6B HOMOLOG, ... | | 著者 | Aufderheide, A, Beck, F, Stengel, F, Hartwig, M, Schweitzer, A, Pfeifer, G, Goldberg, A.L, Sakata, E, Baumeister, W, Foerster, F. | | 登録日 | 2015-06-17 | | 公開日 | 2015-07-22 | | 最終更新日 | 2017-08-30 | | 実験手法 | ELECTRON MICROSCOPY (9.5 Å) | | 主引用文献 | Structural Characterization of the Interaction of Ubp6 with the 26S Proteasome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7NAC
 
 | | State E2 nucleolar 60S ribosomal biogenesis intermediate - Composite model | | 分子名称: | 25S rRNA, 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | 著者 | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | 登録日 | 2021-06-21 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.04 Å) | | 主引用文献 | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7NAD
 
 | | State E2 nucleolar 60S ribosomal biogenesis intermediate - Spb4 local refinement model | | 分子名称: | 25S rRNA, 5.8S rRNA, 60S ribosomal protein L17-A, ... | | 著者 | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | 登録日 | 2021-06-21 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.04 Å) | | 主引用文献 | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7NAF
 
 | | State E2 nucleolar 60S ribosomal biogenesis intermediate - Spb1-MTD local model | | 分子名称: | 25S rRNA, 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | 著者 | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | 登録日 | 2021-06-21 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.13 Å) | | 主引用文献 | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
2GDA
 
 | | REFINED SOLUTION STRUCTURE OF THE GLUCOCORTICOID RECEPTOR DNA-BINDING DOMAIN | | 分子名称: | GLUCOCORTICOID RECEPTOR, ZINC ION | | 著者 | Baumann, H, Paulsen, K, Kovacs, H, Berglund, H, Wright, A.P.H, Gustafsson, J.-A, Hard, T. | | 登録日 | 1994-03-15 | | 公開日 | 1994-06-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Refined solution structure of the glucocorticoid receptor DNA-binding domain.
Biochemistry, 32, 1993
|
|
8AXC
 
 | | Crystal structure of mouse Ces2c | | 分子名称: | Acylcarnitine hydrolase, CHLORIDE ION, NICOTINAMIDE, ... | | 著者 | Eisner, H, Riegler-Berket, L, Rodriguez Gamez, C, Sagmeister, T, Chalhoub, G, Darnhofer, B, Panikkaveetil Jawaharlal, J, Birner-Gruenberger, R, Pavkov-Keller, T, Haemmerle, G, Schoiswohl, G, Oberer, M. | | 登録日 | 2022-08-31 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | The Crystal Structure of Mouse Ces2c, a Potential Ortholog of Human CES2, Shows Structural Similarities in Substrate Regulation and Product Release to Human CES1.
Int J Mol Sci, 23, 2022
|
|
8PD3
 
 | | Ligand-free SpSLC9C1 in lipid nanodiscs, protomer state 2 | | 分子名称: | Sperm-specific sodium proton exchanger | | 著者 | Kalienkova, V, Peter, M, Rheinberger, J, Paulino, C. | | 登録日 | 2023-06-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structures of a sperm-specific solute carrier gated by voltage and cAMP.
Nature, 623, 2023
|
|
