5WYJ
 
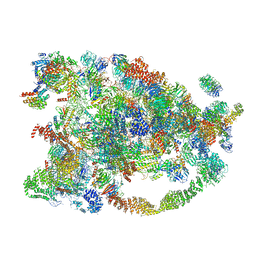 | | Cryo-EM structure of the 90S small subunit pre-ribosome (Dhr1-depleted, Enp1-TAP, state 1) | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, 18S ribosomal RNA, 40S ribosomal protein S1-A, ... | | 著者 | Ye, K, Zhu, X, Sun, Q. | | 登録日 | 2017-01-13 | | 公開日 | 2017-03-29 | | 最終更新日 | 2019-10-09 | | 実験手法 | ELECTRON MICROSCOPY (8.7 Å) | | 主引用文献 | Molecular architecture of the 90S small subunit pre-ribosome.
Elife, 6, 2017
|
|
4UJD
 
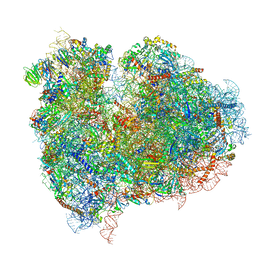 | | mammalian 80S HCV-IRES initiation complex with eIF5B PRE-like state | | 分子名称: | 18S Ribosomal RNA, 28S Ribosomal RNA, 40S RIBOSOMAL PROTEIN ES1, ... | | 著者 | Yamamoto, H, Unbehaun, A, Loerke, J, Behrmann, E, Marianne, C, Burger, J, Mielke, T, Spahn, C.M.T. | | 登録日 | 2014-06-18 | | 公開日 | 2014-07-30 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (8.9 Å) | | 主引用文献 | Structure of the Mammalian 80S Initiation Complex with Eif5B on Hcv Ires
Nat.Struct.Mol.Biol., 21, 2014
|
|
4V4N
 
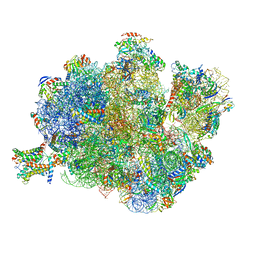 | | Structure of the Methanococcus jannaschii ribosome-SecYEBeta channel complex | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein L7AE, ... | | 著者 | Menetret, J.F, Park, E, Gumbart, J.C, Ludtke, S.J, Li, W, Whynot, A, Rapoport, T.A, Akey, C.W. | | 登録日 | 2013-06-17 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (9 Å) | | 主引用文献 | Structure of the SecY channel during initiation of protein translocation.
Nature, 506, 2013
|
|
4D61
 
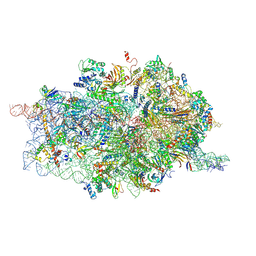 | | Cryo-EM structures of ribosomal 80S complexes with termination factors and cricket paralysis virus IRES reveal the IRES in the translocated state | | 分子名称: | 18S RRNA, 40S RIBOSOMAL PROTEIN S10, 40S RIBOSOMAL PROTEIN S11, ... | | 著者 | Muhs, M, Hilal, T, Mielke, T, Skabkin, M.A, Sanbonmatsu, K.Y, Pestova, T.V, Spahn, C.M.T. | | 登録日 | 2014-11-07 | | 公開日 | 2015-03-04 | | 最終更新日 | 2017-08-30 | | 実験手法 | ELECTRON MICROSCOPY (9 Å) | | 主引用文献 | Cryo-Em of Ribosomal 80S Complexes with Termination Factors Reveals the Translocated Cricket Paralysis Virus Ires.
Mol.Cell, 57, 2015
|
|
4D5L
 
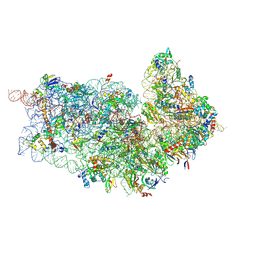 | | Cryo-EM structures of ribosomal 80S complexes with termination factors and cricket paralysis virus IRES reveal the IRES in the translocated state | | 分子名称: | 18S RRNA 2, 40S RIBOSOMAL PROTEIN ES1, 40S RIBOSOMAL PROTEIN ES10, ... | | 著者 | Muhs, M, Hilal, T, Mielke, T, Skabkin, M.A, Sanbonmatsu, K.Y, Pestova, T.V, Spahn, C.M.T. | | 登録日 | 2014-11-05 | | 公開日 | 2015-02-04 | | 最終更新日 | 2017-08-23 | | 実験手法 | ELECTRON MICROSCOPY (9 Å) | | 主引用文献 | Cryo-Em of Ribosomal 80S Complexes with Termination Factors Reveals the Translocated Cricket Paralysis Virus Ires.
Mol.Cell, 57, 2015
|
|
4UJC
 
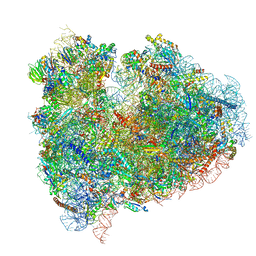 | | mammalian 80S HCV-IRES initiation complex with eIF5B POST-like state | | 分子名称: | 18S RIBOSOMAL RNA, 28S RIBOSOMAL RNA, 40S RIBOSOMAL PROTEIN ES1, ... | | 著者 | Yamamoto, H, Unbehaun, A, Loerke, J, Behrmann, E, Marianne, C, Burger, J, Mielke, T, Spahn, C.M.T. | | 登録日 | 2014-06-18 | | 公開日 | 2014-07-30 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (9.5 Å) | | 主引用文献 | Structure of the Mammalian 80S Initiation Complex with Initiation Factor 5B on Hcv-Ires RNA.
Nat.Struct.Mol.Biol., 21, 2014
|
|
5IDF
 
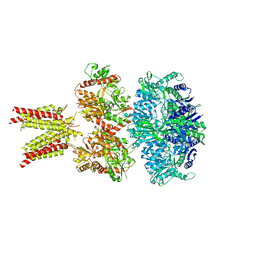 | | Cryo-EM structure of GluA2/3 AMPA receptor heterotetramer (model II) | | 分子名称: | Glutamate receptor 2, Glutamate receptor 3 | | 著者 | Herguedas, B, Garcia-Nafria, J, Fernandez-Leiro, R, Greger, I.H. | | 登録日 | 2016-02-24 | | 公開日 | 2016-03-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (10.31 Å) | | 主引用文献 | Structure and organization of heteromeric AMPA-type glutamate receptors.
Science, 352, 2016
|
|
8P5D
 
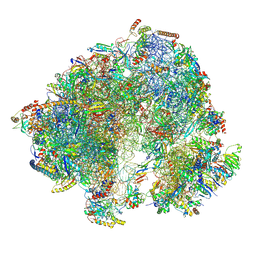 | | Spraguea lophii ribosome in the closed conformation by cryo sub tomogram averaging | | 分子名称: | 40S Ribosomal protein S19, 40S ribosomal protein S0, 40S ribosomal protein S10, ... | | 著者 | Gil Diez, P, McLaren, M, Isupov, M.N, Daum, B, Conners, R, Williams, B. | | 登録日 | 2023-05-23 | | 公開日 | 2023-06-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (10.8 Å) | | 主引用文献 | CryoEM reveals that ribosomes in microsporidian spores are locked in a dimeric hibernating state.
Nat Microbiol, 8, 2023
|
|
7D63
 
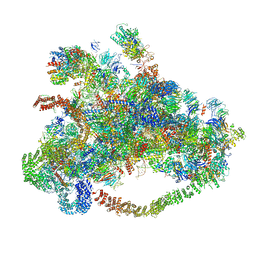 | | Cryo-EM structure of 90S preribosome with inactive Utp24 (state C) | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | 著者 | Du, Y, Zhang, J, An, W, Ye, K. | | 登録日 | 2020-09-29 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (12.3 Å) | | 主引用文献 | Cryo-EM structure of 90S preribosome with inactive Utp24 (state C)
To Be Published
|
|
8P60
 
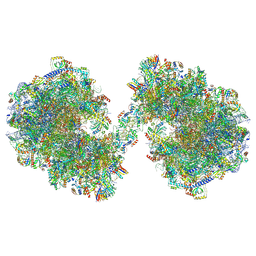 | | Spraguea lophii ribosome dimer | | 分子名称: | 40S Ribosomal protein S19, 40S ribosomal protein S0, 40S ribosomal protein S1, ... | | 著者 | Gil Diez, P, McLaren, M, Isupov, M.N, Daum, B, Conners, R, Williams, B. | | 登録日 | 2023-05-24 | | 公開日 | 2023-06-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (14.3 Å) | | 主引用文献 | CryoEM reveals that ribosomes in microsporidian spores are locked in a dimeric hibernating state.
Nat Microbiol, 8, 2023
|
|
7KTS
 
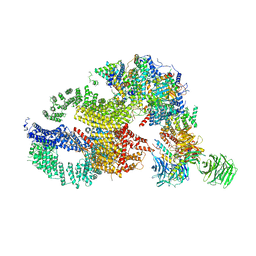 | | Negative stain EM structure of the human SAGA coactivator complex (TRRAP, core, splicing module) | | 分子名称: | Ataxin-7, Isoform 3 of Transcription factor SPT20 homolog, STAGA complex 65 subunit gamma, ... | | 著者 | Herbst, D.A, Esbin, M.N, Nogales, E. | | 登録日 | 2020-11-24 | | 公開日 | 2021-11-10 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (19.09 Å) | | 主引用文献 | Structure of the human SAGA coactivator complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5FKI
 
 | | Pseudorabies virus (PrV) nuclear egress complex proteins fitted as a hexameric lattice into a sub-tomogram average derived from focused- ion beam milled lamellae electron cryo-microscopic data | | 分子名称: | CHLORIDE ION, UL31, UL34 protein, ... | | 著者 | Hagen, C, Dent, K.C, Zeev Ben Mordehai, T, Vasishtan, D, Antonin, W, Mettenleiter, T.C, Gruenewald, K. | | 登録日 | 2015-10-16 | | 公開日 | 2016-03-16 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (35 Å) | | 主引用文献 | Crystal Structure of the Herpesvirus Nuclear Egress Complex Provides Insights Into Inner Nuclear Membrane Remodelling
Cell Rep., 13, 2015
|
|
8CX6
 
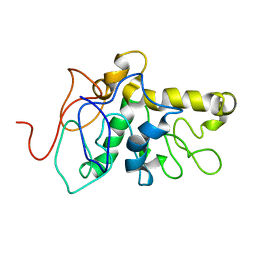 | | TPX2 Minimal Active Domain on Microtubules | | 分子名称: | Targeting protein for Xklp2-A | | 著者 | Guo, C, Alfaro-Aco, R, Russell, R, Zhang, C, Petry, S, Polenova, T. | | 登録日 | 2022-05-19 | | 公開日 | 2023-06-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Structural basis of protein condensation on microtubules underlying branching microtubule nucleation.
Nat Commun, 14, 2023
|
|
7FBR
 
 | |
2LKW
 
 | |
8S9Y
 
 | |
1DV0
 
 | |
5NOC
 
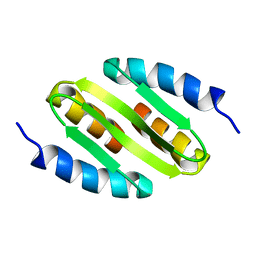 | |
2LZP
 
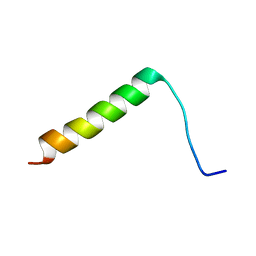 | |
7FBV
 
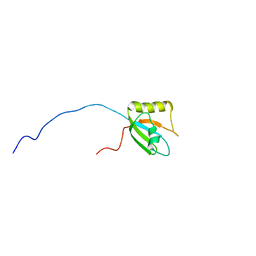 | |
2N68
 
 | |
1F4I
 
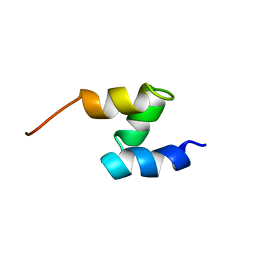 | | SOLUTION STRUCTURE OF THE HHR23A UBA(2) MUTANT P333E, DEFICIENT IN BINDING THE HIV-1 ACCESSORY PROTEIN VPR | | 分子名称: | UV EXCISION REPAIR PROTEIN PROTEIN RAD23 HOMOLOG A | | 著者 | Withers-Ward, E.S, Mueller, T.D, Chen, I.S, Feigon, J. | | 登録日 | 2000-06-07 | | 公開日 | 2000-12-20 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Biochemical and structural analysis of the interaction between the UBA(2) domain of the DNA repair protein HHR23A and HIV-1 Vpr.
Biochemistry, 39, 2000
|
|
1HI7
 
 | |
7CK5
 
 | | Solution structure of 28 amino acid polypeptide (354-381) in Plantago asiatica mosaic virus replicase bound to SDS micelle | | 分子名称: | PlAMV replicase peptide from RNA-dependent RNA polymerase | | 著者 | Komatsu, K, Sasaki, N, Yoshida, T, Suzuki, K, Masujima, Y, Hashimoto, M, Watanabe, S, Tochio, N, Kigawa, T, Yamaji, Y, Oshima, K, Namba, S, Nelson, R, Arie, T. | | 登録日 | 2020-07-15 | | 公開日 | 2021-07-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Identification of a Proline-Kinked Amphipathic alpha-Helix Downstream from the Methyltransferase Domain of a Potexvirus Replicase and Its Role in Virus Replication and Perinuclear Complex Formation.
J.Virol., 95, 2021
|
|
5O1Q
 
 | | LysF1 sh3b domain structure | | 分子名称: | sh3b domain | | 著者 | Benesik, M, Novacek, J, Janda, L, Dopitova, R, Pernisova, M, Melkova, K, Tisakova, L, Doskar, J, Zidek, L, Hejatko, J, Pantucek, R. | | 登録日 | 2017-05-19 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Role of SH3b binding domain in a natural deletion mutant of Kayvirus endolysin LysF1 with a broad range of lytic activity.
Virus Genes, 54, 2018
|
|
