4OML
 
 | |
6L1L
 
 | |
6KZA
 
 | | Crystal structure of the complex of the interaction domains of E. coli DnaB helicase and DnaC helicase loader | | 分子名称: | DNA replication protein DnaC, Replicative DNA helicase | | 著者 | Nagata, K, Okada, A, Ohtsuka, J, Ohkuri, T, Akama, Y, Sakiyama, Y, Miyazaki, E, Horita, S, Katayama, T, Ueda, T, Tanokura, M. | | 登録日 | 2019-09-23 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Crystal structure of the complex of the interaction domains of Escherichia coli DnaB helicase and DnaC helicase loader: structural basis implying a distortion-accumulation mechanism for the DnaB ring opening caused by DnaC binding.
J.Biochem., 167, 2020
|
|
4NXJ
 
 | | Crystal Structure of PF3D7_1475600, a bromodomain from Plasmodium Falciparum | | 分子名称: | Bromodomain protein | | 著者 | Wernimont, A.K, Loppnau, P, Knapp, S, Fonseca, M, Brennan, P.E, Dong, A, Walker, J.R, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Hutchinson, A, Structural Genomics Consortium (SGC) | | 登録日 | 2013-12-09 | | 公開日 | 2014-03-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Crystal Structure of PF3D7_1475600, a bromodomain from Plasmodium Falciparum
TO BE PUBLISHED
|
|
4M8U
 
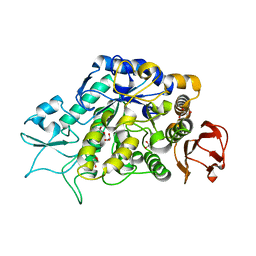 | | The Structure of MalL mutant enzyme V200A from Bacillus subtilus | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | 著者 | Hobbs, J.K, Jiao, W, Easter, A.D, Parker, E.J, Schipper, L.A, Arcus, V.L. | | 登録日 | 2013-08-13 | | 公開日 | 2013-09-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Change in heat capacity for enzyme catalysis determines temperature dependence of enzyme catalyzed rates.
Acs Chem.Biol., 8, 2013
|
|
6DE6
 
 | |
6DPY
 
 | |
6DX9
 
 | |
6E06
 
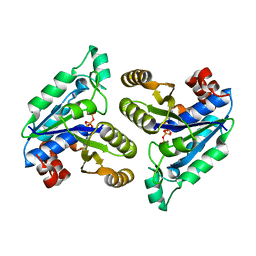 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with cytidine triphosphate solved by precipitant-ligand exchange (crystals grown in citrate precipitant) | | 分子名称: | ATP-dependent dethiobiotin synthetase BioD, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | 著者 | Thompson, A.P, Wegener, K.L, Bruning, J.B, Polyak, S.W. | | 登録日 | 2018-07-06 | | 公開日 | 2018-10-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Precipitant-ligand exchange technique reveals the ADP binding mode in Mycobacterium tuberculosis dethiobiotin synthetase.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6E05
 
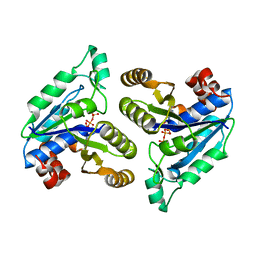 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with cytidine triphosphate solved by precipitant-ligand exchange (crystals grown in sulfate precipitant) | | 分子名称: | ATP-dependent dethiobiotin synthetase BioD, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | 著者 | Thompson, A.P, Wegener, K.L, Bruning, J.B, Polyak, S.W. | | 登録日 | 2018-07-06 | | 公開日 | 2018-10-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Precipitant-ligand exchange technique reveals the ADP binding mode in Mycobacterium tuberculosis dethiobiotin synthetase.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6EA2
 
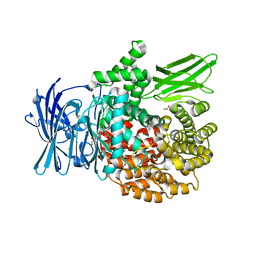 | |
6E6J
 
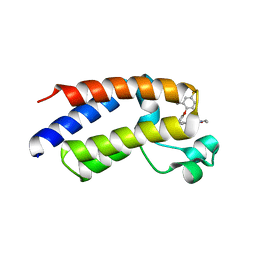 | | BRD2_Bromodomain2 complex with inhibitor 744 | | 分子名称: | Bromodomain-containing protein 2, N-ethyl-4-[2-(4-fluoro-2,6-dimethylphenoxy)-5-(2-hydroxypropan-2-yl)phenyl]-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide | | 著者 | Longenecker, K.L, Park, C.H, Bigelow, L. | | 登録日 | 2018-07-25 | | 公開日 | 2019-07-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Selective inhibition of the BD2 bromodomain of BET proteins in prostate cancer.
Nature, 578, 2020
|
|
6E6S
 
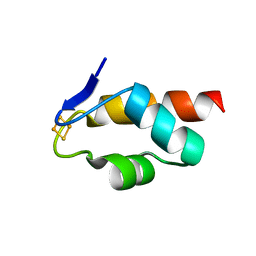 | | 1.45 A resolution structure of the C-terminally truncated [2Fe-2S] ferredoxin (Bfd) R26E/K46Y mutant from Pseudomonas aeruginosa | | 分子名称: | Bacterioferritin-associated ferredoxin, FE2/S2 (INORGANIC) CLUSTER | | 著者 | Lovell, S, Wijerathne, H, Battaile, K.P, Yao, H, Wang, Y, Rivera, M. | | 登録日 | 2018-07-25 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Bfd, a New Class of [2Fe-2S] Protein That Functions in Bacterial Iron Homeostasis, Requires a Structural Anion Binding Site.
Biochemistry, 57, 2018
|
|
6EAA
 
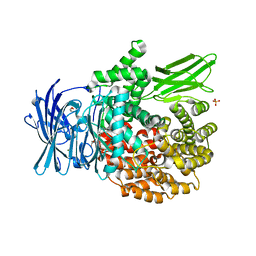 | |
4WEJ
 
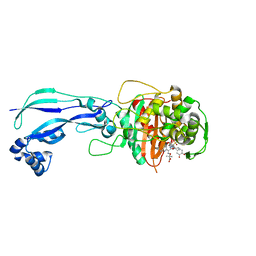 | | Crystal structure of Pseudomonas aeruginosa PBP3 with a R4 substituted allyl monocarbam | | 分子名称: | (3R,4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-4-formyl-1-[({3-[(5R)-5-hydroxy-4-oxo-4,5-dihydropyridin-2-yl]-4-[3-(methylsulfonyl)propyl]-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl}sulfonyl)amino]-10,10-dimethyl-1,6-dioxo-3-(prop-2-en-1-yl)-9-oxa-2,5,8-triazaundec-7-en-11-oic acid, Penicillin-binding protein 3 | | 著者 | Ferguson, A.D. | | 登録日 | 2014-09-10 | | 公開日 | 2015-04-22 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.045 Å) | | 主引用文献 | SAR and Structural Analysis of Siderophore-Conjugated Monocarbam Inhibitors of Pseudomonas aeruginosa PBP3.
Acs Med.Chem.Lett., 6, 2015
|
|
3WS3
 
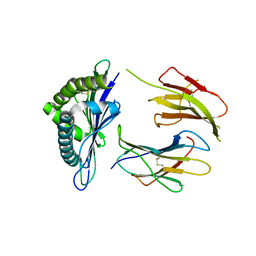 | | Crystal Structure of H-2D in complex with an insulin derived peptide | | 分子名称: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | 著者 | Kumar, P.R, Mukherjee, G, Samanta, D, DiLorenzo, T.P, Almo, S.C, Immune Function Network, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2014-02-28 | | 公開日 | 2014-03-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.335 Å) | | 主引用文献 | Compensatory mechanisms allow undersized anchor-deficient class I MHC ligands to mediate pathogenic autoreactive T cell responses
J. Immunol., 193, 2014
|
|
3RZA
 
 | |
7AAC
 
 | | Crystal structure of the catalytic domain of human PARP1 in complex with veliparib | | 分子名称: | (2R)-2-(7-carbamoyl-1H-benzimidazol-2-yl)-2-methylpyrrolidinium, Poly [ADP-ribose] polymerase 1, SULFATE ION | | 著者 | Schimpl, M, Ogden, T.E.H, Yang, J.-C, Easton, L.E, Underwood, E, Rawlins, P.B, Johannes, J.W, Embrey, K.J, Neuhaus, D. | | 登録日 | 2020-09-04 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.593 Å) | | 主引用文献 | Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acids Res., 49, 2021
|
|
7AAD
 
 | | Crystal structure of the catalytic domain of human PARP1 in complex with olaparib | | 分子名称: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, Poly [ADP-ribose] polymerase 1, SULFATE ION | | 著者 | Schimpl, M, Ogden, T.E.H, Yang, J.-C, Easton, L.E, Underwood, E, Rawlins, P.B, Johannes, J.W, Embrey, K.J, Neuhaus, D. | | 登録日 | 2020-09-04 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acids Res., 49, 2021
|
|
7AAX
 
 | | Crystal structure of MerTK kinase domain in complex with LDC1267 | | 分子名称: | CHLORIDE ION, Tyrosine-protein kinase Mer, ~{N}-[4-(6,7-dimethoxyquinolin-4-yl)oxy-3-fluoranyl-phenyl]-4-ethoxy-1-(4-fluoranyl-2-methyl-phenyl)pyrazole-3-carboxamide | | 著者 | Schimpl, M, Pflug, A, McCoull, W, Nissink, J.W.M, Overman, R.C, Rawlins, P.B, Truman, C, Underwood, E, Warwicker, J, Winter-Holt, J. | | 登録日 | 2020-09-05 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.762 Å) | | 主引用文献 | A-loop interactions in Mer tyrosine kinase give rise to inhibitors with two-step mechanism and long residence time of binding.
Biochem.J., 477, 2020
|
|
7AB0
 
 | | Apo crystal structure of the MerTK kinase domain | | 分子名称: | CHLORIDE ION, Tyrosine-protein kinase Mer | | 著者 | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Overman, R.C, Rawlins, P.B, Truman, C, Underwood, E, Warwicker, J, Winter-Holt, J. | | 登録日 | 2020-09-05 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | A-loop interactions in Mer tyrosine kinase give rise to inhibitors with two-step mechanism and long residence time of binding.
Biochem.J., 477, 2020
|
|
7A8G
 
 | |
7A86
 
 | |
7A7O
 
 | |
7A8O
 
 | |
