5ONI
 
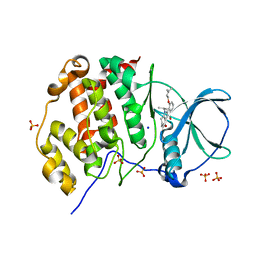 | | LOW-SALT STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA) IN COMPLEX WITH THE INDENOINDOLE-TYPE INHIBITOR 4P | | 分子名称: | 1,4-BUTANEDIOL, 4-(3-methylbut-2-enoxy)-5-propan-2-yl-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, CHLORIDE ION, ... | | 著者 | Hochscherf, J, Lindenblatt, D, Witulski, B, Birus, R, Aichele, D, Marminon, C, Bouaziz, Z, Le Borgne, M, Jose, J, Niefind, K. | | 登録日 | 2017-08-03 | | 公開日 | 2017-12-27 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Unexpected Binding Mode of a Potent Indeno[1,2-b]indole-Type Inhibitor of Protein Kinase CK2 Revealed by Complex Structures with the Catalytic Subunit CK2 alpha and Its Paralog CK2 alpha '.
Pharmaceuticals (Basel), 10, 2017
|
|
6V0L
 
 | | PDGFR-b Promoter Forms a G-Vacancy Quadruplex that Can be Complemented by dGMP: Molecular Structure and Recognition of Guanine Derivatives and Metabolites | | 分子名称: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, DNA (5'-D(*(3D1)P*AP*GP*GP*GP*AP*GP*GP*GP*CP*GP*GP*CP*GP*GP*GP*AP*CP*A)-3') | | 著者 | Wang, K.B, Dickerhoff, J, Wu, G, Yang, D. | | 登録日 | 2019-11-18 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | PDGFR-beta Promoter Forms a Vacancy G-Quadruplex that Can Be Filled in by dGMP: Solution Structure and Molecular Recognition of Guanine Metabolites and Drugs.
J.Am.Chem.Soc., 142, 2020
|
|
1BJA
 
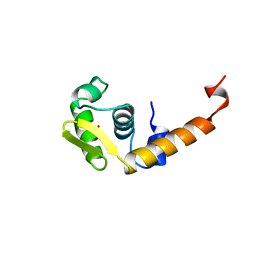 | | ACTIVATION DOMAIN OF THE PHAGE T4 TRANSCRIPTION FACTOR MOTA | | 分子名称: | SULFATE ION, TRANSCRIPTION REGULATORY PROTEIN MOTA | | 著者 | Finnin, M.S, Cicero, M.P, Davies, C, Porter, S.J, White, S.W, Kreuzer, K.N. | | 登録日 | 1998-06-23 | | 公開日 | 1998-11-04 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | The activation domain of the MotA transcription factor from bacteriophage T4.
EMBO J., 16, 1997
|
|
4H22
 
 | |
8FVI
 
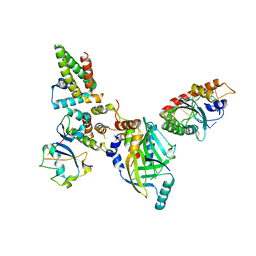 | | Human APOBEC3H bound to HIV-1 Vif in complex with CBF-beta, ELOB, ELOC, and CUL5 | | 分子名称: | Core-binding factor subunit beta, Cullin 5, DNA dC->dU-editing enzyme APOBEC-3H, ... | | 著者 | Ito, F, Alvarez-Cabrera, A.L, Zhou, Z.H, Chen, X.S. | | 登録日 | 2023-01-19 | | 公開日 | 2023-09-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.24 Å) | | 主引用文献 | Structural basis of HIV-1 Vif-mediated E3 ligase targeting of host APOBEC3H.
Nat Commun, 14, 2023
|
|
6TGT
 
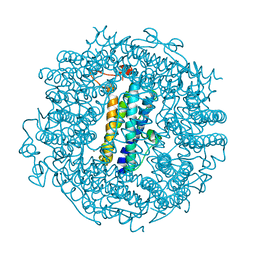 | |
7OUF
 
 | | Structure of the STLV intasome:B56 complex bound to the strand-transfer inhibitor XZ450 | | 分子名称: | 4-azanyl-~{N}-[[2,4-bis(fluoranyl)phenyl]methyl]-6-[3-(dimethylamino)-3-oxidanylidene-propyl]-1-oxidanyl-2-oxidanylidene-1,8-naphthyridine-3-carboxamide, DNA (5'-D(*AP*CP*TP*GP*TP*GP*TP*TP*TP*GP*GP*CP*GP*CP*TP*TP*CP*TP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*AP*GP*AP*AP*GP*CP*GP*CP*CP*AP*AP*AP*CP*AP*CP*A)-3'), ... | | 著者 | Barski, M.S, Ballandras-Colas, A, Cronin, N.B, Pye, V.E, Cherepanov, P, Maertens, G.N. | | 登録日 | 2021-06-11 | | 公開日 | 2021-08-18 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for the inhibition of HTLV-1 integration inferred from cryo-EM deltaretroviral intasome structures.
Nat Commun, 12, 2021
|
|
3JVS
 
 | | Characterization of the Chk1 allosteric inhibitor binding site | | 分子名称: | 2-[(4-tert-butyl-3-nitrophenyl)carbonyl]-N-naphthalen-1-ylhydrazinecarboxamide, Serine/threonine-protein kinase Chk1 | | 著者 | Chen, P. | | 登録日 | 2009-09-17 | | 公開日 | 2009-10-06 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Characterization of the CHK1 allosteric inhibitor binding site.
Biochemistry, 48, 2009
|
|
3JVR
 
 | |
8RZB
 
 | | IL-1beta in complex with covalent DEL hit | | 分子名称: | 8-[4-methyl-3-(trifluoromethyl)phenyl]-2-[[(7S)-7-(2-morpholin-4-ylethylcarbamoyl)-4-(phenylsulfonyl)-1,4-diazepan-1-yl]carbonyl]imidazo[1,2-a]pyridine-6-carboxylic acid, Interleukin-1 beta | | 著者 | Rondeau, J.-M, Lehmann, S. | | 登録日 | 2024-02-12 | | 公開日 | 2024-05-22 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.836 Å) | | 主引用文献 | Ligandability Assessment of IL-1 beta by Integrated Hit Identification Approaches.
J.Med.Chem., 67, 2024
|
|
1AO2
 
 | | cobalt(III)-deglycopepleomycin determined by NMR studies | | 分子名称: | AGLYCON OF PEPLOMYCIN, COBALT (III) ION, HYDROGEN PEROXIDE | | 著者 | Caceres-Cortes, J, Sugiyama, H, Ikudome, K, Saito, I, Wang, A.H.-J. | | 登録日 | 1997-07-16 | | 公開日 | 1999-07-30 | | 最終更新日 | 2024-03-13 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structures of cobalt(III)-pepleomycin and cobalt(III)-deglycopepleomycin (green forms) determined by NMR studies.
Eur.J.Biochem., 244, 1997
|
|
8GRM
 
 | | Cryo-EM structure of PRC1 bound to H2AK119-UbcH5b-Ub nucleosome | | 分子名称: | COMMD3 protein, DNA (144-MER), DNA (145-MER), ... | | 著者 | Ai, H.S, Zebin, T, Zhihend, D, Jiakun, T, Liying, Z, Jia-Bin, L, Man, P, Liu, L. | | 登録日 | 2022-09-02 | | 公開日 | 2023-04-12 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Synthetic E2-Ub-nucleosome conjugates for studying nucleosome ubiquitination.
Chem, 2023
|
|
8GJ5
 
 | | fungal pcna and peptidomimetic | | 分子名称: | Proliferating cell nuclear antigen, THR-ASP-ILE-ARG-ASN-PHE-PHE-HIS-SER | | 著者 | Vandborg, B, Bruning, J.B. | | 登録日 | 2023-03-14 | | 公開日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Towards a High-Affinity Peptidomimetic Targeting Proliferating Cell Nuclear Antigen from Aspergillus fumigatus.
J Fungi, 9, 2023
|
|
1KXS
 
 | |
6RVS
 
 | | Atomic structure of the Epstein-Barr portal, structure II | | 分子名称: | Portal protein | | 著者 | Machon, C, Fabrega-Ferrer, M, Zhou, D, Cuervo, A, Carrascosa, J.L, Stuart, D.I, Coll, M. | | 登録日 | 2019-05-31 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.59 Å) | | 主引用文献 | Atomic structure of the Epstein-Barr virus portal.
Nat Commun, 10, 2019
|
|
5WH1
 
 | |
1AO4
 
 | | COBALT(III)-PEPLOMYCIN COMPLEX DETERMINED BY NMR STUDIES | | 分子名称: | 3-O-carbamoyl-alpha-D-mannopyranose-(1-2)-alpha-L-gulopyranose, AGLYCON OF PEPLOMYCIN, COBALT (III) ION, ... | | 著者 | Caceres-Cortes, J, Sugiyama, H, Ikudome, K, Saito, I, Wang, A.H.-J. | | 登録日 | 1997-07-16 | | 公開日 | 1999-07-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structures of cobalt(III)-pepleomycin and cobalt(III)-deglycopepleomycin (green forms) determined by NMR studies.
Eur.J.Biochem., 244, 1997
|
|
3ABU
 
 | | Crystal Structure of LSD1 in complex with a 2-PCPA derivative, S1201 | | 分子名称: | Lysine-specific histone demethylase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-5-[(1R,3R,3aS)-3-[2-(benzyloxy)-3-fluorophenyl]-1-hydroxy-10,11-dimethyl-4,6-dioxo-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate | | 著者 | Mimasu, S, Umezawa, N, Sato, S, Higuchi, T, Umehara, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2009-12-21 | | 公開日 | 2010-07-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structurally Designed trans-2-Phenylcyclopropylamine Derivatives Potently Inhibit Histone Demethylase LSD1/KDM1
Biochemistry, 49, 2010
|
|
3ABT
 
 | | Crystal Structure of LSD1 in complex with trans-2-pentafluorophenylcyclopropylamine | | 分子名称: | Lysine-specific histone demethylase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-2,3,4-trihydroxy-5-[(1R,3R,3aS)-1-hydroxy-10,11-dimethyl-4,6-dioxo-3-(pentafluorophenyl)-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]pentyl dihydrogen diphosphate | | 著者 | Mimasu, S, Umezawa, N, Sato, S, Higuchi, T, Umehara, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2009-12-21 | | 公開日 | 2010-07-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structurally Designed trans-2-Phenylcyclopropylamine Derivatives Potently Inhibit Histone Demethylase LSD1/KDM1
Biochemistry, 49, 2010
|
|
4Z1V
 
 | | Structure of Factor Inhibiting HIF (FIH) in complex with Fe, NO, and NOG | | 分子名称: | DI(HYDROXYETHYL)ETHER, FE (III) ION, Hypoxia-inducible factor 1-alpha inhibitor, ... | | 著者 | Taabazuing, C.Y, Garman, S.C, Knapp, M.J. | | 登録日 | 2015-03-27 | | 公開日 | 2016-01-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Substrate Promotes Productive Gas Binding in the alpha-Ketoglutarate-Dependent Oxygenase FIH.
Biochemistry, 55, 2016
|
|
6H2X
 
 | | MukB coiled-coil elbow from E. coli | | 分子名称: | Chromosome partition protein MukB,Chromosome partition protein MukB | | 著者 | Buermann, F, Lowe, J. | | 登録日 | 2018-07-16 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A folded conformation of MukBEF and cohesin.
Nat.Struct.Mol.Biol., 26, 2019
|
|
4N06
 
 | |
2W9R
 
 | | Structural basis of N-end rule substrate recognition in Escherichia coli by the ClpAP adaptor protein ClpS | | 分子名称: | ATP-DEPENDENT CLP PROTEASE ADAPTER PROTEIN CLPS, DNA PROTECTION DURING STARVATION PROTEIN | | 著者 | Schuenemann, V, Kralik, S.M, Albrecht, R, Spall, S.K, Truscott, K.N, Dougan, D.A, Zeth, K. | | 登録日 | 2009-01-28 | | 公開日 | 2009-04-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural Basis of N-End Rule Substrate Recognition in Escherichia Coli by the Clpap Adaptor Protein Clps.
Embo Rep., 10, 2009
|
|
3WKJ
 
 | | The nucleosome containing human TSH2B | | 分子名称: | CHLORIDE ION, DNA (145-MER), Histone H2A type 1-B/E, ... | | 著者 | Urahama, T, Horikoshi, N, Osakabe, A, Tachiwana, H, Kurumizaka, H. | | 登録日 | 2013-10-22 | | 公開日 | 2014-04-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of human nucleosome containing the testis-specific histone variant TSH2B.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
5D7U
 
 | | Crystal structure of the C-terminal domain of MMTV integrase | | 分子名称: | ISOPROPYL ALCOHOL, Pr160 | | 著者 | Cook, N.J, Pye, V.E, Ballandras-Colas, A, Engelman, A, Cherepanov, P. | | 登録日 | 2015-08-14 | | 公開日 | 2016-02-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function.
Nature, 530, 2016
|
|
