7CN2
 
 | |
8OOR
 
 | | CryoEM Structure INO80core Hexasome complex Rvb core refinement state2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-05 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.87 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OW4
 
 | | 2.75 angstrom crystal structure of human NFAT1 with bound DNA | | 分子名称: | DNA (5'-D(*AP*AP*CP*TP*AP*TP*TP*TP*TP*TP*CP*CP*AP*GP*C)-3'), DNA (5'-D(*TP*TP*GP*CP*TP*GP*GP*AP*AP*AP*AP*AP*TP*AP*G)-3'), Nuclear factor of activated T-cells, ... | | 著者 | Lopez-Sagaseta, J, Erausquin, E, Hernandez-Morales, S, Urdiciain, A, Lasarte, J.J, Lozano, T. | | 登録日 | 2023-04-27 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | 2.75 angstrom crystal structure of human NFAT1 with bound DNA
Not published
|
|
7CFM
 
 | | Cryo-EM structure of the P395-bound GPBAR-Gs complex | | 分子名称: | 2-(ethylamino)-6-[3-(4-propan-2-ylphenyl)propanoyl]-7,8-dihydro-5H-pyrido[4,3-d]pyrimidine-4-carboxamide, CHOLESTEROL, G-protein coupled bile acid receptor 1, ... | | 著者 | Yang, F, Mao, C, Guo, L, Lin, J, Ming, Q, Xiao, P, Wu, X, Shen, Q, Guo, S, Shen, D, Lu, R, Zhang, L, Huang, S, Ping, Y, Zhang, C, Ma, C, Zhang, K, Liang, X, Shen, Y, Nan, F, Yi, F, Luca, V, Zhou, J, Jiang, C, Sun, J, Xie, X, Yu, X, Zhang, Y. | | 登録日 | 2020-06-27 | | 公開日 | 2020-09-09 | | 最終更新日 | 2020-12-02 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis of GPBAR activation and bile acid recognition.
Nature, 587, 2020
|
|
8OO9
 
 | | CryoEM Structure INO80core Hexasome complex ATPase-DNA refinement state1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Chromatin-remodeling ATPase INO80, DNA strand 1, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-04 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOA
 
 | | CryoEM Structure INO80core Hexasome complex Hexasome refinement state1 | | 分子名称: | DNA Strand 2, DNA strand 1, Histone H2A, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-04 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOP
 
 | | CryoEM Structure INO80core Hexasome complex composite model state2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-05 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOS
 
 | | CryoEM Structure INO80core Hexasome complex ATPase-hexasome refinement state 2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Chromatin-remodeling ATPase Ino80, DNA Strand 2, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-05 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8P2B
 
 | | Crystal structure of CbFMN4 domain 1 | | 分子名称: | Clostridiaceae bacterium FMN4 domain 1, FLAVIN MONONUCLEOTIDE | | 著者 | Rozeboom, H.J, Fraaije, M.W. | | 登録日 | 2023-05-15 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Characterization of two bacterial multi-flavinylated proteins harboring multiple covalent flavin cofactors.
Bba Adv, 4, 2023
|
|
8ONQ
 
 | |
8OOC
 
 | | CryoEM Structure INO80core Hexasome complex Rvb core refinement state1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Chromatin-remodeling ATPase Ino80, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-05 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OMU
 
 | |
8OKR
 
 | |
8P2K
 
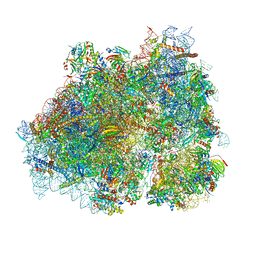 | | Ternary complex of translating ribosome, NAC and METAP1 | | 分子名称: | 18s rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | 著者 | Jia, M, Jaskolowski, M, Scaiola, A, Jomaa, A, Ban, N. | | 登録日 | 2023-05-16 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | NAC controls cotranslational N-terminal methionine excision in eukaryotes.
Science, 380, 2023
|
|
8P4Y
 
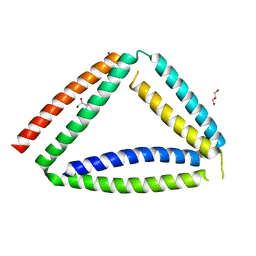 | |
8OVW
 
 | | Cryo-EM structure of CBF1-CCAN bound topologically to centromeric DNA | | 分子名称: | C0N3 DNA, Centromere-binding protein 1, Inner kinetochore subunit AME1, ... | | 著者 | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | 登録日 | 2023-04-26 | | 公開日 | 2023-08-09 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
7CAH
 
 | | The interface of H014 Fab binds to SARS-CoV-2 S | | 分子名称: | Heavy chain of H014 Fab, Light chain of H014 Fab, Spike protein S1 | | 著者 | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Rao, Z, wang, Y, Qin, C, Wang, X. | | 登録日 | 2020-06-08 | | 公開日 | 2020-08-12 | | 最終更新日 | 2021-03-10 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
8OW0
 
 | | Cryo-EM structure of CBF1-CCAN bound topologically to a centromeric CENP-A nucleosome | | 分子名称: | C0N3 DNA, Centromere-binding protein 1, Histone H2A.1, ... | | 著者 | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | 登録日 | 2023-04-26 | | 公開日 | 2023-08-09 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
8OLT
 
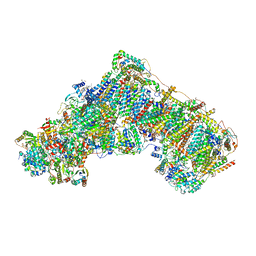 | | Mitochondrial complex I from Mus musculus in the active state bound with piericidin A | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Grba, D.N, Chung, I, Bridges, H.R, Agip, A.N.A, Hirst, J. | | 登録日 | 2023-03-30 | | 公開日 | 2023-08-09 | | 実験手法 | ELECTRON MICROSCOPY (2.84 Å) | | 主引用文献 | Investigation of hydrated channels and proton pathways in a high-resolution cryo-EM structure of mammalian complex I.
Sci Adv, 9, 2023
|
|
8OO6
 
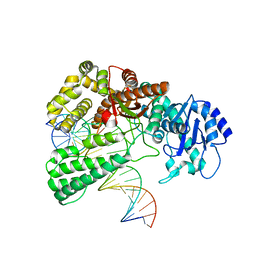 | | Pol I bound to extended and displaced DNA section - closed conformation | | 分子名称: | DNA polymerase I, Displaced primer, Extending Primer, ... | | 著者 | Botto, M, Borsellini, A, Lamers, M.H. | | 登録日 | 2023-04-04 | | 公開日 | 2023-08-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | A four-point molecular handover during Okazaki maturation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8EV2
 
 | | Dual Modulators | | 分子名称: | (3aS,4R,9bR)-4-(2-chloro-4-hydroxyphenyl)-2,3,3a,4,5,9b-hexahydro-1H-cyclopenta[c]quinoline-8-sulfonamide, (3~{a}~{R},4~{S},9~{b}~{S})-4-(2-chloranyl-4-oxidanyl-phenyl)-2,3,3~{a},4,5,9~{b}-hexahydro-1~{H}-cyclopenta[c]quinoline-8-sulfonamide, Estrogen receptor, ... | | 著者 | Tinivella, A, Nwachukwu, J.C, Angeli, A, Foschi, F, Benatti, A.L, Pinzi, L, Izard, T, Ferraroni, M, Rangarajan, E.S, Christodoulou, M, Passarella, D, Supuran, C, Nettles, K.W, Rastelli, G. | | 登録日 | 2022-10-19 | | 公開日 | 2022-12-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Design, synthesis, biological evaluation and crystal structure determination of dual modulators of carbonic anhydrases and estrogen receptors.
Eur.J.Med.Chem., 246, 2022
|
|
8OSD
 
 | |
7CRP
 
 | | NSD3 bearing E1181K/T1232A dual mutation in complex with 187-bp NCP (1:1 binding mode) | | 分子名称: | DNA (168-MER), Histone H2A, Histone H2B, ... | | 著者 | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | 登録日 | 2020-08-14 | | 公開日 | 2020-10-21 | | 最終更新日 | 2021-03-03 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
4W8B
 
 | | Crystal structure of XEG5B, a GH5 xyloglucan-specific beta-1,4-glucanase from ruminal metagenomic library, in complex with XXLG | | 分子名称: | Exo-xyloglucanase, GLYCEROL, SULFATE ION, ... | | 著者 | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | 登録日 | 2014-08-22 | | 公開日 | 2015-03-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
8P3V
 
 | | Homomeric GluA1 in tandem with TARP gamma-3, desensitized conformation 3 | | 分子名称: | Glutamate receptor 1 flip isoform, Voltage-dependent calcium channel gamma-3 subunit | | 著者 | Zhang, D, Krieger, J.M, Yamashita, K, Greger, I.H. | | 登録日 | 2023-05-18 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3.53 Å) | | 主引用文献 | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
