6ZW2
 
 | | Crystal structure of OXA-10loop48 in complex with hydrolyzed meropenem | | 分子名称: | (2S,3R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfan yl-3-methyl-2,3-dihydro-1H-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | 著者 | Tassone, G, Di Pisa, F, Benvenuti, M, De Luca, F, Pozzi, C, Mangani, S, Docquier, J.D. | | 登録日 | 2020-07-27 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Mechanistic insights into carbapenem hydrolysis by OXA-48 and the OXA10-derived hybrids OXA-10 loop24 and loop48
To Be Published
|
|
6ZRP
 
 | | Crystal structure of class D Beta-lactamase OXA-48 in complex with meropenem | | 分子名称: | (2S,3R,4S)-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | 著者 | Tassone, G, De Luca, F, Pozzi, C, Di Pisa, F, Benvenuti, M, Mangani, S, Doquier, J.D. | | 登録日 | 2020-07-14 | | 公開日 | 2021-07-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Mechanistic insights into carbapenem hydrolysis by OXA-48 and the OXA10-derived hybrids OXA-10 loop24 and loop48
To Be Published
|
|
6ZRJ
 
 | | Crystal structure of class D Beta-lactamase OXA-48 in complex with ertapenem | | 分子名称: | (2S,3R,4S)-4-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-2-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | 著者 | Tassone, G, Di Pisa, F, Benvenuti, M, De Luca, F, Pozzi, C, Docquier, J.D, Mangani, S. | | 登録日 | 2020-07-13 | | 公開日 | 2021-07-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Mechanistic insights into carbapenem hydrolysis by OXA-48 and the OXA10-derived hybrids OXA-10 loop24 and loop48
To Be Published
|
|
6ZRI
 
 | | Crystal structure of OXA-10loop24 in complex with meropenem | | 分子名称: | (2S,3R,4S)-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase | | 著者 | Tassone, G, Di Pisa, F, Benvenuti, M, De Luca, F, Pozzi, C, Mangani, S, Docquier, J.D. | | 登録日 | 2020-07-13 | | 公開日 | 2021-07-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Mechanistic insights into carbapenem hydrolysis by OXA-48 and the OXA10-derived hybrids OXA-10 loop24 and loop48
To Be Published
|
|
6ZRH
 
 | | Crystal structure of OXA-10loop24 in complex with ertapenem | | 分子名称: | (2S,3R,4S)-4-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-2-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase | | 著者 | Tassone, G, Di Pisa, F, Benvenuti, M, De Luca, F, Pozzi, C, Mangani, S, Docquier, J.D. | | 登録日 | 2020-07-13 | | 公開日 | 2021-07-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Mechanistic insights into carbapenem hydrolysis by OXA-48 and the OXA10-derived hybrids OXA-10 loop24 and loop48
To Be Published
|
|
6ZRG
 
 | | Crystal structure of OXA-10loop48 in complex with hydrolyzed doripenem | | 分子名称: | (4R,5S)-3-{[(3S,5S)-5-({[amino(dihydroxy)methyl]amino}methyl)pyrrolidin-3-yl]sulfanyl}-5-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | 著者 | Tassone, G, Di Pisa, F, Benvenuti, M, De Luca, F, Pozzi, C, Docquier, J.D, Mangani, S. | | 登録日 | 2020-07-13 | | 公開日 | 2021-07-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Mechanistic insights into carbapenem hydrolysis by OXA-48 and the OXA10-derived hybrids OXA-10 loop24 and loop48
To Be Published
|
|
6YN0
 
 | | Structure of E. coli PBP1b with a FtsN peptide activating transglycosylase activity | | 分子名称: | Cell division protein FtsN, MOENOMYCIN, Penicillin-binding protein 1B | | 著者 | Kerff, F, Terrak, M, Boes, A, Herman, H, Charlier, P. | | 登録日 | 2020-04-10 | | 公開日 | 2020-11-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The bacterial cell division protein fragment E FtsN binds to and activates the major peptidoglycan synthase PBP1b.
J.Biol.Chem., 295, 2020
|
|
6Y6Z
 
 | | Structure of Pseudomonas aeruginosa Penicillin-Binding Protein 3 (PBP3) in complex with Compound 1 | | 分子名称: | GLYCEROL, Peptidoglycan D,D-transpeptidase FtsI, ~{tert}-butyl ~{N}-[(2~{S})-2-methyl-4-oxidanyl-1-oxidanylidene-pent-4-en-2-yl]carbamate | | 著者 | Newman, H, Bellini, D, Dowson, C.G. | | 登録日 | 2020-02-27 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Demonstration of the utility of DOS-derived fragment libraries for rapid hit derivatisation in a multidirectional fashion.
Chem Sci, 11, 2020
|
|
6Y6U
 
 | | Structure of Pseudomonas aeruginosa Penicillin-Binding Protein 3 (PBP3) in complex with Compound 6 | | 分子名称: | 2-(4-hydroxyphenyl)-~{N}-[(2~{S})-2-methyl-4-oxidanyl-1-oxidanylidene-pent-4-en-2-yl]ethanamide, GLYCEROL, Peptidoglycan D,D-transpeptidase FtsI | | 著者 | Newman, H, Bellini, D, Dowson, C.G. | | 登録日 | 2020-02-27 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Demonstration of the utility of DOS-derived fragment libraries for rapid hit derivatisation in a multidirectional fashion
Chem Sci, 11, 2020
|
|
6XV5
 
 | |
6XQZ
 
 | | Crystal structure of the catalytic domain of PBP2 S310A from Neisseria gonorrhoeae at pH 7.5 | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptidoglycan D,D-transpeptidase PenA, ... | | 著者 | Fenton, B.A, Zhou, P, Davies, C. | | 登録日 | 2020-07-10 | | 公開日 | 2021-07-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Mutations in PBP2 from ceftriaxone-resistant Neisseria gonorrhoeae alter the dynamics of the beta 3-beta 4 loop to favor a low-affinity drug-binding state.
J.Biol.Chem., 297, 2021
|
|
6XQY
 
 | |
6XQX
 
 | | Crystal structure of the catalytic domain of PBP2 S310A from Neisseria gonorrhoeae with the H514A mutation at pH 7.5 | | 分子名称: | 1,2-ETHANEDIOL, Probable peptidoglycan D,D-transpeptidase PenA, SULFATE ION | | 著者 | Fenton, B.A, Zhou, P, Davies, C. | | 登録日 | 2020-07-10 | | 公開日 | 2021-07-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Mutations in PBP2 from ceftriaxone-resistant Neisseria gonorrhoeae alter the dynamics of the beta 3-beta 4 loop to favor a low-affinity drug-binding state.
J.Biol.Chem., 297, 2021
|
|
6XQV
 
 | | Crystal structure of the catalytic domain of PBP2 S310A from Neisseria gonorrhoeae in a pre-acylation complex with ceftriaxone | | 分子名称: | CHLORIDE ION, Ceftriaxone, Probable peptidoglycan D,D-transpeptidase PenA, ... | | 著者 | Fenton, B.A, Zhou, P, Davies, C. | | 登録日 | 2020-07-10 | | 公開日 | 2021-07-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Mutations in PBP2 from ceftriaxone-resistant Neisseria gonorrhoeae alter the dynamics of the beta 3-beta 4 loop to favor a low-affinity drug-binding state.
J.Biol.Chem., 297, 2021
|
|
6XQR
 
 | | OXA-48 bound by Compound 2.2 | | 分子名称: | Beta-lactamase, CHLORIDE ION, [1,1'-biphenyl]-4,4'-disulfonic acid | | 著者 | Taylor, D.M, Hu, L, Prasad, B.V.V, Palzkill, T. | | 登録日 | 2020-07-10 | | 公開日 | 2021-12-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
6XJ3
 
 | | Crystal structure of Class D beta-lactamase from Klebsiella quasipneumoniae in complex with avibactam | | 分子名称: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-06-22 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Class D beta-lactamase from Klebsiella quasipneumoniae
To Be Published
|
|
6WY4
 
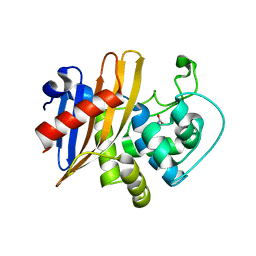 | | Crystal Structure of Wild Type Class D beta-lactamase from Clostridium difficile 630 | | 分子名称: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SODIUM ION | | 著者 | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-05-12 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of Wild Type Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
6WHL
 
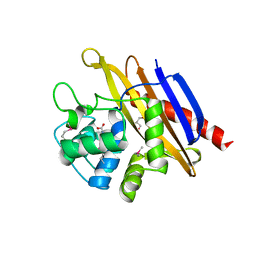 | |
6W5O
 
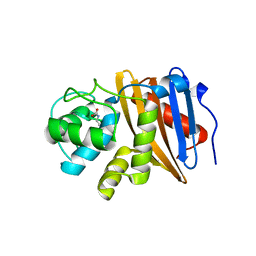 | | Class D beta-lactamase BAT-2 delta mutant | | 分子名称: | 1,2-ETHANEDIOL, BAT-2 Beta-lactamase delta mutant, CITRATE ANION | | 著者 | Smith, C.A, Vakulenko, S.B, Stewart, N.K, Toth, M. | | 登録日 | 2020-03-13 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | A surface loop modulates activity of the Bacillus class D beta-lactamases.
J.Struct.Biol., 211, 2020
|
|
6W5G
 
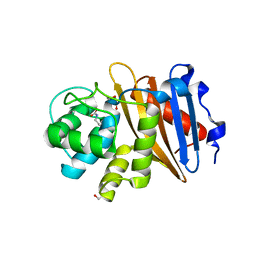 | | Class D beta-lactamase BAT-2 | | 分子名称: | 1,2-ETHANEDIOL, BAT-2 beta-lactamase | | 著者 | Smith, C.A, Vakulenko, S.B, Stewart, N.K, Toth, M. | | 登録日 | 2020-03-13 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.451 Å) | | 主引用文献 | A surface loop modulates activity of the Bacillus class D beta-lactamases.
J.Struct.Biol., 211, 2020
|
|
6W5F
 
 | | Class D beta-lactamase BSU-2 delta mutant | | 分子名称: | 1,2-ETHANEDIOL, BSU-2delta mutant, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Smith, C.A, Vakulenko, S.B, Stewart, N.K, Toth, M. | | 登録日 | 2020-03-13 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A surface loop modulates activity of the Bacillus class D beta-lactamases.
J.Struct.Biol., 211, 2020
|
|
6W5E
 
 | | Class D beta-lactamase BSU-2 | | 分子名称: | 1,2-ETHANEDIOL, BSU-2 beta-lactamase, MALONATE ION | | 著者 | Smith, C.A, Vakulenko, S.B, Stewart, N.K, Toth, M. | | 登録日 | 2020-03-13 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | A surface loop modulates activity of the Bacillus class D beta-lactamases.
J.Struct.Biol., 211, 2020
|
|
6VOT
 
 | | Crystal structure of Pseudomonas aerugonisa PBP3 complexed to gamma-lactam YU253434 | | 分子名称: | 1-[(2S)-2-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}-3-oxopropyl]-4-{[2-(5,6 -dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl]carbamoyl}-2,5-dihydro-1H-pyrazole-3-carboxylic acid, Peptidoglycan D,D-transpeptidase FtsI | | 著者 | van den Akker, F. | | 登録日 | 2020-01-31 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | A gamma-Lactam Siderophore Antibiotic Effective against Multidrug-Resistant Gram-Negative Bacilli.
J.Med.Chem., 63, 2020
|
|
6VJE
 
 | | Crystal structure of Pseudomonas aeruginosa penicillin-binding protein 3 (PBP3) complexed with ceftobiprole | | 分子名称: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, CHLORIDE ION, Peptidoglycan D,D-transpeptidase FtsI | | 著者 | van den Akker, F, Kumar, V. | | 登録日 | 2020-01-15 | | 公開日 | 2020-03-11 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Structural Insights into Ceftobiprole Inhibition of Pseudomonas aeruginosa Penicillin-Binding Protein 3.
Antimicrob.Agents Chemother., 64, 2020
|
|
6VBM
 
 | | Crystal structure of a S310A mutant of PBP2 from Neisseria gonorrhoeae | | 分子名称: | PHOSPHATE ION, Probable peptidoglycan D,D-transpeptidase PenA | | 著者 | Singh, A, Davies, C. | | 登録日 | 2019-12-19 | | 公開日 | 2020-04-15 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Mutations in Neisseria gonorrhoeae penicillin-binding protein 2 associated with extended-spectrum cephalosporin resistance create an energetic barrier against acylation via restriction of protein dynamics
J.Biol.Chem., 2020
|
|
