4V8I
 
 | | Crystal structure of YfiA bound to the 70S ribosome. | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | 著者 | Polikanov, Y.S, Blaha, G.M, Steitz, T.A. | | 登録日 | 2011-12-12 | | 公開日 | 2014-07-09 | | 最終更新日 | 2014-12-10 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | How hibernation factors RMF, HPF, and YfiA turn off protein synthesis.
Science, 336, 2012
|
|
4V9R
 
 | | Crystal structure of antibiotic DITYROMYCIN bound to 70S ribosome | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | 著者 | Bulkley, D.P, Brandi, L, Polikanov, Y.S, Fabbretti, A, O'Connor, M, Gualerzi, C.O, Steitz, T.A. | | 登録日 | 2013-12-05 | | 公開日 | 2014-07-09 | | 最終更新日 | 2014-12-10 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The antibiotics dityromycin and GE82832 bind protein S12 and block EF-G-catalyzed translocation.
Cell Rep, 6, 2014
|
|
4V6G
 
 | | Initiation complex of 70S ribosome with two tRNAs and mRNA. | | 分子名称: | 16S RRNA (E.COLI NUMBERING), 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | 著者 | Jenner, L.B, Yusupova, G, Yusupov, M. | | 登録日 | 2009-07-10 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structural aspects of messenger RNA reading frame maintenance by the ribosome.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6GSL
 
 | | Structure of T. thermophilus 70S ribosome complex with mRNA, tRNAfMet and cognate tRNAArg in the A-site | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Rozov, A, Yusupov, M, Yusupova, G. | | 登録日 | 2018-06-14 | | 公開日 | 2018-07-04 | | 最終更新日 | 2018-09-05 | | 実験手法 | X-RAY DIFFRACTION (3.16 Å) | | 主引用文献 | Tautomeric G•U pairs within the molecular ribosomal grip and fidelity of decoding in bacteria.
Nucleic Acids Res., 46, 2018
|
|
7NS3
 
 | | Substrate receptor scaffolding module of yeast Chelator-GID SR4 E3 ubiquitin ligase bound to Fbp1 substrate | | 分子名称: | BJ4_G0018240.mRNA.1.CDS.1, Fructose-bisphosphatase, Glucose-induced degradation protein 8, ... | | 著者 | Sherpa, D, Chrustowicz, J, Prabu, J.R, Schulman, B.A. | | 登録日 | 2021-03-05 | | 公開日 | 2021-05-05 | | 最終更新日 | 2021-07-07 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme.
Mol.Cell, 81, 2021
|
|
7VGW
 
 | | Yeast gid10 with Pro-peptide | | 分子名称: | BJ4_G0041530.mRNA.1.CDS.1 | | 著者 | Shin, J.S, Park, S.H, Kim, L, Heo, J, Song, H.K. | | 登録日 | 2021-09-19 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of yeast Gid10 in complex with Pro/N-degron.
Biochem.Biophys.Res.Commun., 582, 2021
|
|
7BP3
 
 | | Cryo-EM structure of the human MCT2 | | 分子名称: | Monocarboxylate transporter 2 | | 著者 | Zhang, B, Jin, Q, Zhang, X, Guo, J, Ye, S. | | 登録日 | 2020-03-21 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cooperative transport mechanism of human monocarboxylate transporter 2.
Nat Commun, 11, 2020
|
|
7AN4
 
 | |
7ANG
 
 | |
7ANC
 
 | |
4NXT
 
 | | Crystal structure of the cytosolic domain of human MiD51 | | 分子名称: | GLYCEROL, Mitochondrial dynamic protein MID51, SULFATE ION | | 著者 | Richter, V, Ryan, M.T, Kvansakul, M. | | 登録日 | 2013-12-09 | | 公開日 | 2013-12-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Structural and functional analysis of MiD51, a dynamin receptor required for mitochondrial fission.
J.Cell Biol., 204, 2014
|
|
4NXV
 
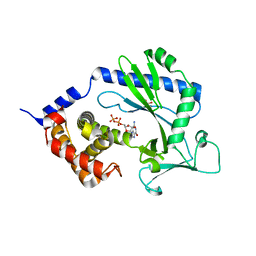 | | Crystal structure of the cytosolic domain of human MiD51 | | 分子名称: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, Mitochondrial dynamic protein MID51, ... | | 著者 | Richter, V, Kvansakul, M, Ryan, M.T. | | 登録日 | 2013-12-09 | | 公開日 | 2013-12-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and functional analysis of MiD51, a dynamin receptor required for mitochondrial fission.
J.Cell Biol., 204, 2014
|
|
4NXU
 
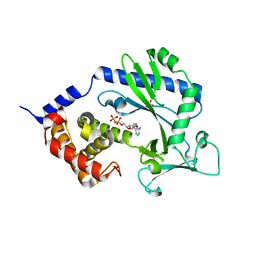 | | Crystal structure of the cytosolic domain of human MiD51 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, Mitochondrial dynamic protein MID51, ... | | 著者 | Richter, V, Kvansakul, M, Ryan, M.T. | | 登録日 | 2013-12-09 | | 公開日 | 2013-12-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and functional analysis of MiD51, a dynamin receptor required for mitochondrial fission.
J.Cell Biol., 204, 2014
|
|
4NXX
 
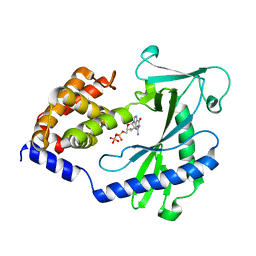 | |
4NXW
 
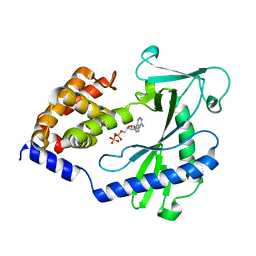 | |
2MAM
 
 | |
2YRV
 
 | | Solution structure of the RBB1NT domain of human RB(retinoblastoma)-binding protein 1 | | 分子名称: | AT-rich interactive domain-containing protein 4A | | 著者 | Nameki, N, Saito, K, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-04-03 | | 公開日 | 2008-04-08 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the RBB1NT domain of human RB(retinoblastoma)-binding protein 1
To be Published
|
|
7QQY
 
 | |
5V8T
 
 | | Crystal structure of SMT fusion Peptidyl-prolyl cis-trans isomerase from Burkholderia pseudomallei complexed with SF354 | | 分子名称: | 2-{[3,5-bis(2-methoxyethoxy)benzene-1-carbonyl]amino}ethyl (2S)-1-(benzylsulfonyl)piperidine-2-carboxylate, FORMIC ACID, MAGNESIUM ION, ... | | 著者 | Lorimer, D.D, Dranow, D.M, Seufert, F, Abendroth, J, Holzgrabe, U. | | 登録日 | 2017-03-22 | | 公開日 | 2018-03-14 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of SMT fusion Peptidyl-prolyl cis-trans isomerase from Burkholderia pseudomallei complexed with SF354
to be published
|
|
5VDC
 
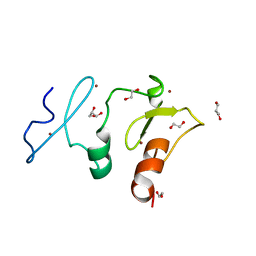 | | Crystal structure of the human DPF2 tandem PHD finger domain | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, ZINC ION, ... | | 著者 | Huber, F.M, Davenport, A.M, Hoelz, A. | | 登録日 | 2017-04-01 | | 公開日 | 2017-05-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Histone-binding of DPF2 mediates its repressive role in myeloid differentiation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7WUG
 
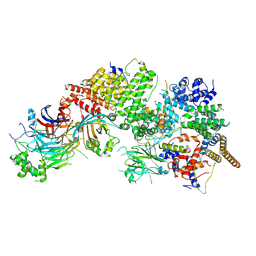 | | GID subcomplex: Gid12 bound Substrate Receptor Scaffolding module | | 分子名称: | Glucose-induced degradation protein 8, HLJ1_G0042170.mRNA.1.CDS.1, Vacuolar import and degradation protein 24, ... | | 著者 | Qiao, S, Cheng, J.D, Schulman, B.A. | | 登録日 | 2022-02-08 | | 公開日 | 2022-06-08 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM structures of Gid12-bound GID E3 reveal steric blockade as a mechanism inhibiting substrate ubiquitylation.
Nat Commun, 13, 2022
|
|
6O49
 
 | |
6O4A
 
 | |
4FAO
 
 | | Specificity and Structure of a high affinity Activin-like 1 (ALK1) signaling complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-2B, Growth/differentiation factor 2, ... | | 著者 | Townson, S.A, Martinez-Hackert, E, Greppi, C, Lowden, P, Sako, D, Liu, J, Ucran, J.A, Liharska, K, Underwood, K.W, Seehra, J, Kumar, R, Grinberg, A.V. | | 登録日 | 2012-05-22 | | 公開日 | 2012-06-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.357 Å) | | 主引用文献 | Specificity and Structure of a High Affinity Activin Receptor-like Kinase 1 (ALK1) Signaling Complex.
J.Biol.Chem., 287, 2012
|
|
4OAF
 
 | |
