3M2R
 
 | | Structural Insight into Methyl-Coenzyme M Reductase Chemistry using Coenzyme B Analogues | | 分子名称: | 1,2-ETHANEDIOL, 1-THIOETHANESULFONIC ACID, Coenzyme B, ... | | 著者 | Cedervall, P.E, Dey, M, Ragsdale, S.W, Wilmot, C.M. | | 登録日 | 2010-03-08 | | 公開日 | 2010-09-15 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structural insight into methyl-coenzyme M reductase chemistry using coenzyme B analogues.
Biochemistry, 49, 2010
|
|
3MGQ
 
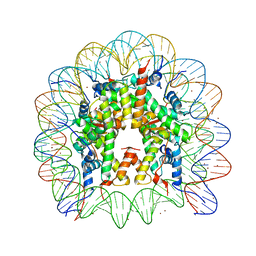 | | Binding of Nickel ions to the Nucleosome Core Particle | | 分子名称: | CHLORIDE ION, DNA (147-MER), Histone H2A, ... | | 著者 | Mohideen, K, Muhammad, R, Davey, C.A. | | 登録日 | 2010-04-07 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Perturbations in nucleosome structure from heavy metal association.
Nucleic Acids Res., 38, 2010
|
|
3MB3
 
 | | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) | | 分子名称: | 1-methylpyrrolidin-2-one, PH-interacting protein | | 著者 | Filippakopoulos, P, Picaud, S, Keates, T, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2010-03-25 | | 公開日 | 2010-04-14 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
4INU
 
 | | Yeast 20S proteasome in complex with the vinyl sulfone LU112 | | 分子名称: | N3Phe-Phe(4-NH2CH2)-Leu-Phe(4-NH2CH2)-methyl vinyl sulfone, bound form, Proteasome component C1, ... | | 著者 | Geurink, P.P, van der Linden, W.A, Mirabella, A.C, Gallastegui, N, de Bruin, G, Blom, A.E.M, Voges, M.J, Mock, E.D, Florea, B.I, van der Marel, G.A, Driessen, C, van der Stelt, M, Groll, M, Overkleeft, H.S, Kisselev, A.F. | | 登録日 | 2013-01-06 | | 公開日 | 2013-01-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Incorporation of Non-natural Amino Acids Improves Cell Permeability and Potency of Specific Inhibitors of Proteasome Trypsin-like Sites.
J.Med.Chem., 56, 2013
|
|
1ZA7
 
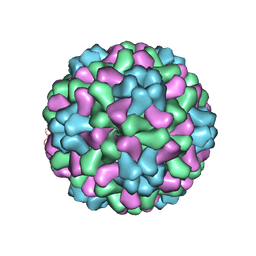 | | The crystal structure of salt stable cowpea cholorotic mottle virus at 2.7 angstroms resolution. | | 分子名称: | Coat protein | | 著者 | Bothner, B, Speir, J.A, Qu, C, Willits, D.A, Young, M.J, Johnson, J.E. | | 登録日 | 2005-04-05 | | 公開日 | 2006-03-21 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Enhanced local symmetry interactions globally stabilize a mutant virus capsid that maintains infectivity and capsid dynamics.
J.Virol., 80, 2006
|
|
7B75
 
 | | Cryo-EM Structure of Human Thyroglobulin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Thyroglobulin, ... | | 著者 | Adaixo, R, Righetto, R, Steiner, E.M, Taylor, N.M.I, Stahlberg, H. | | 登録日 | 2020-12-09 | | 公開日 | 2021-12-29 | | 最終更新日 | 2022-01-26 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structure of native human thyroglobulin.
Nat Commun, 13, 2022
|
|
3MG8
 
 | | Structure of yeast 20S open-gate proteasome with Compound 16 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N-(2,2-dimethylpropyl)-N~2~-[1H-indol-3-yl(oxo)acetyl]-L-asparaginyl-N-(2-methylbenzyl)-3-pyridin-4-yl-L-alaninamide, ... | | 著者 | Sintchak, M.D. | | 登録日 | 2010-04-05 | | 公開日 | 2011-05-18 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Characterization of a new series of non-covalent proteasome inhibitors with exquisite potency and selectivity for the 20S beta5-subunit.
Biochem.J., 430, 2010
|
|
1ZF2
 
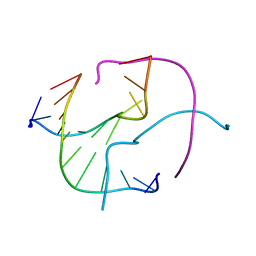 | | Four-stranded DNA Holliday Junction (CCC) | | 分子名称: | 5'-D(*CP*CP*GP*GP*GP*CP*CP*CP*GP*G)-3' | | 著者 | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | 登録日 | 2005-04-19 | | 公開日 | 2005-05-10 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZI9
 
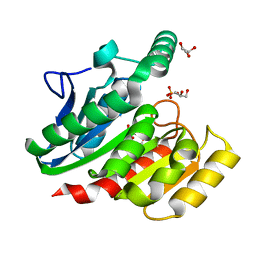 | | Crystal Structure Analysis of the dienelactone hydrolase (E36D, C123S) mutant- 1.5 A | | 分子名称: | Carboxymethylenebutenolidase, GLYCEROL, SULFATE ION | | 著者 | Kim, H.-K, Liu, J.-W, Carr, P.D, Ollis, D.L. | | 登録日 | 2005-04-27 | | 公開日 | 2005-07-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Following directed evolution with crystallography: structural changes observed in changing the substrate specificity of dienelactone hydrolase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ZGN
 
 | | Crystal Structure of the Glutathione Transferase Pi in Complex with Dinitrosyl-diglutathionyl Iron Complex | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FE (III) ION, GLUTATHIONE, ... | | 著者 | Parker, L.J, Adams, J.J, Parker, M.W. | | 登録日 | 2005-04-21 | | 公開日 | 2005-11-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Nitrosylation of human glutathione transferase P1-1 with dinitrosyl diglutathionyl iron complex in vitro and in vivo
J.Biol.Chem., 280, 2005
|
|
5Q1J
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A in complex with FMSOA000341b | | 分子名称: | DNA cross-link repair 1A protein, MALONATE ION, N-(2-hydroxyphenyl)acetamide, ... | | 著者 | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | 登録日 | 2017-05-15 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | PanDDA analysis group deposition
To Be Published
|
|
3MNN
 
 | | A Ruthenium Antitumour Agent Forms Specific Histone Protein Adducts in the Nucleosome Core | | 分子名称: | 1,3,5-triaza-7-phosphatricyclo[3.3.1.1~3,7~]decane, 1-methyl-4-(1-methylethyl)benzene, DNA (145-MER), ... | | 著者 | Ong, M.S, Davey, C.A. | | 登録日 | 2010-04-22 | | 公開日 | 2011-04-06 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A ruthenium antimetastasis agent forms specific histone protein adducts in the nucleosome core
Chemistry, 17, 2011
|
|
3M1V
 
 | | Structural Insight into Methyl-Coenzyme M Reductase Chemistry using Coenzyme B Analogues | | 分子名称: | 1,2-ETHANEDIOL, 1-THIOETHANESULFONIC ACID, ACETATE ION, ... | | 著者 | Cedervall, P.E, Dey, M, Ragsdale, S.W, Wilmot, C.M. | | 登録日 | 2010-03-05 | | 公開日 | 2010-09-15 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structural insight into methyl-coenzyme M reductase chemistry using coenzyme B analogues.
Biochemistry, 49, 2010
|
|
4H24
 
 | | Cytochrome P450BM3-CIS cyclopropanation catalyst | | 分子名称: | Cytochrome P450-BM3 variant P450BM3-Cis, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Coelho, P.S, Wang, Z.J, Ener, M.E, Baril, S.A, Kannan, A, Arnold, F.H, Brustad, E.M. | | 登録日 | 2012-09-11 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A serine-substituted P450 catalyzes highly efficient carbene transfer to olefins in vivo.
Nat.Chem.Biol., 9, 2013
|
|
1ZOA
 
 | | Crystal Structure Of A328V Mutant Of The Heme Domain Of P450Bm-3 With N-Palmitoylglycine | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450:NADPH-P450 reductase, GLYCEROL, ... | | 著者 | Hegda, A, Chen, B, Haines, D.C, Bondlela, M, Mullin, D, Graham, S.E, Tomchick, D.R, Machius, M, Peterson, J.A. | | 登録日 | 2005-05-12 | | 公開日 | 2006-08-01 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | A single active-site mutation of P450BM-3 dramatically enhances substrate binding and rate of product formation.
Biochemistry, 50, 2011
|
|
5NIQ
 
 | |
5NAM
 
 | |
3HYE
 
 | | Crystal structure of 20S proteasome in complex with hydroxylated salinosporamide | | 分子名称: | (2R,3S,4R)-2-[(S)-(1S)-cyclohex-2-en-1-yl(hydroxy)methyl]-3-hydroxy-4-(2-hydroxyethyl)-3-methyl-5-oxopyrrolidine-2-carbaldehyde, Proteasome component C1, Proteasome component C11, ... | | 著者 | Groll, M, Arthur, K.A.M, Macherla, V.R, Manam, R.R, Potts, B.C. | | 登録日 | 2009-06-22 | | 公開日 | 2009-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Snapshots of the fluorosalinosporamide/20S complex offer mechanistic insights for fine tuning proteasome inhibition
J.Med.Chem., 52, 2009
|
|
5PO2
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N10132a | | 分子名称: | 1,2-ETHANEDIOL, 5-hydroxy-1,3-dihydro-2H-indol-2-one, Bromodomain-containing protein 1, ... | | 著者 | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | 登録日 | 2017-02-07 | | 公開日 | 2017-03-15 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
3HXO
 
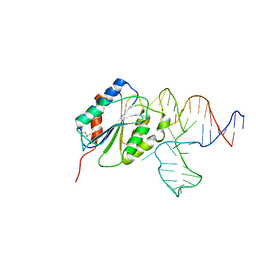 | | Crystal Structure of Von Willebrand Factor (VWF) A1 Domain in Complex with DNA Aptamer ARC1172, an Inhibitor of VWF-Platelet Binding | | 分子名称: | Aptamer ARC1172, von Willebrand factor | | 著者 | Huang, R.H, Sadler, J.E, Fremont, D.H, Diener, J.L, Schaub, R.G. | | 登録日 | 2009-06-21 | | 公開日 | 2009-11-17 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | A structural explanation for the antithrombotic activity of ARC1172, a DNA aptamer that binds von Willebrand factor domain A1.
Structure, 17, 2009
|
|
7B69
 
 | |
7B6C
 
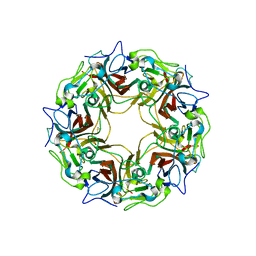 | | BK Polyomavirus VP1 pentamer fusion with long C-terminal extended arm | | 分子名称: | CALCIUM ION, DIMETHYL SULFOXIDE, Major capsid protein VP1,Major capsid protein VP1 | | 著者 | Osipov, E.M, Beelen, S, Strelkov, S.V. | | 登録日 | 2020-12-07 | | 公開日 | 2022-06-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.484 Å) | | 主引用文献 | Discovery of novel druggable pockets on polyomavirus VP1 through crystallographic fragment-based screening to develop capsid assembly inhibitors.
Rsc Chem Biol, 3, 2022
|
|
4OSV
 
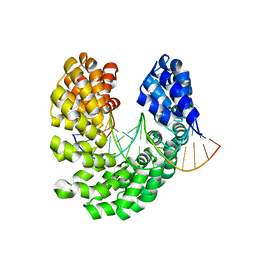 | | Crystal structure of the S505M mutant of TAL effector dHax3 | | 分子名称: | DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*TP*AP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*TP*AP*TP*CP*TP*CP*TP*CP*T)-3'), Hax3 | | 著者 | Deng, D, Wu, J.P, Yan, C.Y, Pan, X.J, Yan, N. | | 登録日 | 2014-02-13 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.996 Å) | | 主引用文献 | Revisiting the TALE repeat
Protein Cell, 5, 2014
|
|
1ZF6
 
 | | TGG DUPLEX A-DNA | | 分子名称: | 5'-D(*CP*CP*CP*CP*AP*TP*GP*GP*GP*G)-3', CALCIUM ION, SODIUM ION | | 著者 | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | 登録日 | 2005-04-19 | | 公開日 | 2005-05-10 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
5NZS
 
 | | The structure of the COPI coat leaf in complex with the ArfGAP2 uncoating factor | | 分子名称: | ADP-ribosylation factor 1, ADP-ribosylation factor GTPase-activating protein 2, Coatomer subunit alpha, ... | | 著者 | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | 登録日 | 2017-05-15 | | 公開日 | 2017-06-28 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (10.1 Å) | | 主引用文献 | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
